Please be patient as the page loads
|
DNM3A_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DNM3A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
DNM3A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987655 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
DNM3B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.973346 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
DNM3B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.973003 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88509, O88510, O88511 | Gene names | Dnmt3b | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase MmuIIIB) (DNA MTase MmuIIIB) (M.MmuIIIB). | |||||
|
DNM3L_HUMAN
|
||||||
| θ value | 5.00389e-67 (rank : 5) | NC score | 0.903424 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UJW3, Q9BUJ4 | Gene names | DNMT3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA (cytosine-5)-methyltransferase 3-like. | |||||
|
DNM3L_MOUSE
|
||||||
| θ value | 4.38363e-63 (rank : 6) | NC score | 0.894756 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CWR8 | Gene names | Dnmt3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA (cytosine-5)-methyltransferase 3-like. | |||||
|
PWWP2_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 7) | NC score | 0.467719 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 8) | NC score | 0.167123 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 9) | NC score | 0.170949 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 10) | NC score | 0.032100 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.148808 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.043555 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TERF2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.067802 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |
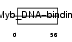
|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.087679 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.050490 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.085970 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.084082 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.086091 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.103333 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
TOIP2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.055370 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
MSH6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.094313 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.017967 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.031298 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SP1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.007711 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08047, Q86TN8, Q9H3Q5, Q9NR51, Q9NY21, Q9NYE7 | Gene names | SP1, TSFP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.061804 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.053086 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
HPS4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.038174 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
SP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.007371 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 730 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O89090, O89087, Q62251, Q64167 | Gene names | Sp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.054444 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ZBT45_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.008098 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |
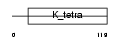
|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.085009 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.029813 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GP179_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.037957 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.088399 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.020069 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
BNIPL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.020218 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z465, Q6DK43, Q8TCY7, Q8WYG2 | Gene names | BNIPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.005853 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
TERF2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.053859 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
GP124_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.012704 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.025222 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.032624 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
HDGR3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.074195 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
HDGR3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.074820 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.086243 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.032063 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.088410 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.022036 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.031625 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.028200 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SP110_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.069071 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.076994 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.076114 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.015735 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
GANAB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.012010 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.020551 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.028330 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.061874 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.068819 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.036626 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FOXK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.005871 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |
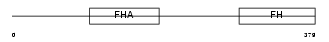
|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
KS6B2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | -0.000555 (rank : 106) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1M4 | Gene names | Rps6kb2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K-beta 2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (S6K2). | |||||
|
P53_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.014250 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |
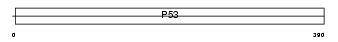
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.019806 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
WDR46_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.025969 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |
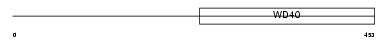
|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.032786 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
K1543_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.021816 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
LY10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.040279 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.029777 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.010826 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
OVOL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.001642 (rank : 105) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTJ2, Q545S2 | Gene names | Ovol1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative transcription factor Ovo-like 1 (mOvo1) (mOvo1a). | |||||
|
PCP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.017683 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IVA1, Q3KRG7 | Gene names | PCP2 | |||
|
Domain Architecture |
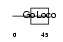
|
|||||
| Description | Purkinje cell protein 2 homolog. | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.011269 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.017706 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.020779 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ANR25_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.005280 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.016614 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
AT1B4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.013985 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99ME6, Q99ME5 | Gene names | Atp1b4 | |||
|
Domain Architecture |
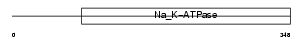
|
|||||
| Description | X/potassium-transporting ATPase subunit beta-m (X,K-ATPase beta-m subunit). | |||||
|
CCD96_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.014495 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.008722 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.059564 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.019696 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
SH2D3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.007942 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.019750 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
THAP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.011989 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WTV1, Q569K1, Q5TH66, Q5TH67, Q8N8T6, Q9BSC7, Q9Y3H2, Q9Y3H3 | Gene names | THAP3 | |||
|
Domain Architecture |
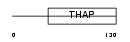
|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
WDR81_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.012972 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5ND34, Q3TA90, Q3TUS9, Q6KAR1 | Gene names | Wdr81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
ATS12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.002330 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.015095 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.016434 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
GEMI5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.005277 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.016969 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
OSBL5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.005528 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
PI51C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.011908 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PPR3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.010094 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.016904 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.021944 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.021883 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.022754 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TBC12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.006457 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.024967 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TRI29_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.005984 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
TRPS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.007293 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
TSSC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.013154 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5U2, Q86VL2, Q9BRS6 | Gene names | TSSC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4 (Tumor-suppressing subchromosomal transferable fragment candidate gene 4 protein) (Tumor-suppressing STF cDNA 4 protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.006618 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ZO3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.004034 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
HDGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.073689 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
HDGF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.053853 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
DNM3A_MOUSE
|
||||||
| NC score | 0.987655 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O88508, Q3UH24, Q8CJ60, Q922J0, Q9CSE1 | Gene names | Dnmt3a | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase MmuIIIA) (DNA MTase MmuIIIA) (M.MmuIIIA). | |||||
|
DNM3B_HUMAN
|
||||||
| NC score | 0.973346 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
DNM3B_MOUSE
|
||||||
| NC score | 0.973003 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88509, O88510, O88511 | Gene names | Dnmt3b | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase MmuIIIB) (DNA MTase MmuIIIB) (M.MmuIIIB). | |||||
|
DNM3L_HUMAN
|
||||||
| NC score | 0.903424 (rank : 5) | θ value | 5.00389e-67 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UJW3, Q9BUJ4 | Gene names | DNMT3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA (cytosine-5)-methyltransferase 3-like. | |||||
|
DNM3L_MOUSE
|
||||||
| NC score | 0.894756 (rank : 6) | θ value | 4.38363e-63 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CWR8 | Gene names | Dnmt3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA (cytosine-5)-methyltransferase 3-like. | |||||
|
PWWP2_HUMAN
|
||||||
| NC score | 0.467719 (rank : 7) | θ value | 1.133e-10 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6NUJ5, Q5SZI0, Q6ZQX5, Q96F43 | Gene names | PWWP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PWWP domain-containing protein 2. | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.170949 (rank : 8) | θ value | 0.00035302 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.167123 (rank : 9) | θ value | 0.00035302 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.148808 (rank : 10) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.103333 (rank : 11) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_HUMAN
|
||||||
| NC score | 0.094313 (rank : 12) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.088410 (rank : 13) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.088399 (rank : 14) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.087679 (rank : 15) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.086243 (rank : 16) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.086091 (rank : 17) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.085970 (rank : 18) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.085009 (rank : 19) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.084082 (rank : 20) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.076994 (rank : 21) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.076114 (rank : 22) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
HDGR3_MOUSE
|
||||||
| NC score | 0.074820 (rank : 23) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JMG7, Q3TRX2, Q8BQ69, Q8BR62, Q9D2M7 | Gene names | Hdgfrp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3). | |||||
|
HDGR3_HUMAN
|
||||||
| NC score | 0.074195 (rank : 24) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y3E1 | Gene names | HDGFRP3, HDGF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatoma-derived growth factor-related protein 3 (HRP-3) (Hepatoma- derived growth factor 2). | |||||
|
HDGF_HUMAN
|
||||||
| NC score | 0.073689 (rank : 25) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51858, Q5SZ09 | Gene names | HDGF, HMG1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF) (High-mobility group protein 1- like 2) (HMG-1L2). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.069071 (rank : 26) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.068819 (rank : 27) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
TERF2_HUMAN
|
||||||
| NC score | 0.067802 (rank : 28) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.061874 (rank : 29) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.061804 (rank : 30) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.059564 (rank : 31) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
TOIP2_MOUSE
|
||||||
| NC score | 0.055370 (rank : 32) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BYU6, Q6NY10, Q8BJP0 | Gene names | Tor1aip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.054444 (rank : 33) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TERF2_MOUSE
|
||||||
| NC score | 0.053859 (rank : 34) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
HDGF_MOUSE
|
||||||
| NC score | 0.053853 (rank : 35) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51859, Q8BPG7, Q9CYA4, Q9JK87 | Gene names | Hdgf, Tdrm1 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatoma-derived growth factor (HDGF). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.053086 (rank : 36) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.050490 (rank : 37) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.043555 (rank : 38) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.040279 (rank : 39) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
HPS4_MOUSE
|
||||||
| NC score | 0.038174 (rank : 40) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.037957 (rank : 41) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.036626 (rank : 42) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.032786 (rank : 43) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.032624 (rank : 44) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.032100 (rank : 45) | θ value | 0.00035302 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.032063 (rank : 46) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.031625 (rank : 47) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.031298 (rank : 48) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.029813 (rank : 49) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.029777 (rank : 50) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.028330 (rank : 51) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.028200 (rank : 52) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
WDR46_MOUSE
|
||||||
| NC score | 0.025969 (rank : 53) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z0H1 | Gene names | Wdr46, Bing4 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 46 (WD repeat protein BING4). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.025222 (rank : 54) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.024967 (rank : 55) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.022754 (rank : 56) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.022036 (rank : 57) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.021944 (rank : 58) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.021883 (rank : 59) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.021816 (rank : 60) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.020779 (rank : 61) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.020551 (rank : 62) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
BNIPL_HUMAN
|
||||||
| NC score | 0.020218 (rank : 63) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z465, Q6DK43, Q8TCY7, Q8WYG2 | Gene names | BNIPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2/adenovirus E1B 19 kDa-interacting protein 2-like protein. | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.020069 (rank : 64) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.019806 (rank : 65) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.019750 (rank : 66) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.019696 (rank : 67) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.017967 (rank : 68) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.017706 (rank : 69) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
PCP2_HUMAN
|
||||||
| NC score | 0.017683 (rank : 70) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IVA1, Q3KRG7 | Gene names | PCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Purkinje cell protein 2 homolog. | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.016969 (rank : 71) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.016904 (rank : 72) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.016614 (rank : 73) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.016434 (rank : 74) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.015735 (rank : 75) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.015095 (rank : 76) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.014495 (rank : 77) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
P53_HUMAN
|
||||||
| NC score | 0.014250 (rank : 78) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
AT1B4_MOUSE
|
||||||
| NC score | 0.013985 (rank : 79) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99ME6, Q99ME5 | Gene names | Atp1b4 | |||
|
Domain Architecture |

|
|||||
| Description | X/potassium-transporting ATPase subunit beta-m (X,K-ATPase beta-m subunit). | |||||
|
TSSC4_HUMAN
|
||||||
| NC score | 0.013154 (rank : 80) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5U2, Q86VL2, Q9BRS6 | Gene names | TSSC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4 (Tumor-suppressing subchromosomal transferable fragment candidate gene 4 protein) (Tumor-suppressing STF cDNA 4 protein). | |||||
|
WDR81_MOUSE
|
||||||
| NC score | 0.012972 (rank : 81) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5ND34, Q3TA90, Q3TUS9, Q6KAR1 | Gene names | Wdr81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.012704 (rank : 82) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
GANAB_HUMAN
|
||||||
| NC score | 0.012010 (rank : 83) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
|
THAP3_HUMAN
|
||||||
| NC score | 0.011989 (rank : 84) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WTV1, Q569K1, Q5TH66, Q5TH67, Q8N8T6, Q9BSC7, Q9Y3H2, Q9Y3H3 | Gene names | THAP3 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 3. | |||||
|
PI51C_HUMAN
|
||||||
| NC score | 0.011908 (rank : 85) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60331, Q7LE07 | Gene names | PIP5K1C, KIAA0589 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type I gamma) (PtdIns(4)P- 5-kinase gamma) (PtdInsPKIgamma) (PIP5KIgamma). | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.011269 (rank : 86) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.010826 (rank : 87) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
PPR3A_MOUSE
|
||||||
| NC score | 0.010094 (rank : 88) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR9, Q8BUJ4, Q8BUL0 | Gene names | Ppp1r3a, Pp1g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 3A (Protein phosphatase 1 glycogen-associated regulatory subunit) (Protein phosphatase type-1 glycogen targeting subunit) (RGL). | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.008722 (rank : 89) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
ZBT45_HUMAN
|
||||||
| NC score | 0.008098 (rank : 90) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
SH2D3_MOUSE
|
||||||
| NC score | 0.007942 (rank : 91) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
SP1_HUMAN
|
||||||
| NC score | 0.007711 (rank : 92) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08047, Q86TN8, Q9H3Q5, Q9NR51, Q9NY21, Q9NYE7 | Gene names | SP1, TSFP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
SP1_MOUSE
|
||||||
| NC score | 0.007371 (rank : 93) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 730 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O89090, O89087, Q62251, Q64167 | Gene names | Sp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp1. | |||||
|
TRPS1_MOUSE
|
||||||
| NC score | 0.007293 (rank : 94) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q925H1 | Gene names | Trps1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.006618 (rank : 95) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
TBC12_HUMAN
|
||||||
| NC score | 0.006457 (rank : 96) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
TRI29_HUMAN
|
||||||
| NC score | 0.005984 (rank : 97) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14134, Q96AA9, Q9BZY7 | Gene names | TRIM29, ATDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 29 (Ataxia-telangiectasia group D- associated protein). | |||||
|
FOXK1_MOUSE
|
||||||
| NC score | 0.005871 (rank : 98) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.005853 (rank : 99) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
OSBL5_MOUSE
|
||||||
| NC score | 0.005528 (rank : 100) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
ANR25_HUMAN
|
||||||
| NC score | 0.005280 (rank : 101) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
GEMI5_MOUSE
|
||||||
| NC score | 0.005277 (rank : 102) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.004034 (rank : 103) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ATS12_HUMAN
|
||||||
| NC score | 0.002330 (rank : 104) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58397, Q6UWL3 | Gene names | ADAMTS12 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 12) (ADAM-TS 12) (ADAM-TS12). | |||||
|
OVOL1_MOUSE
|
||||||
| NC score | 0.001642 (rank : 105) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTJ2, Q545S2 | Gene names | Ovol1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative transcription factor Ovo-like 1 (mOvo1) (mOvo1a). | |||||
|
KS6B2_MOUSE
|
||||||
| NC score | -0.000555 (rank : 106) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1M4 | Gene names | Rps6kb2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase beta-2 (EC 2.7.11.1) (S6K-beta 2) (70 kDa ribosomal protein S6 kinase 2) (p70-S6KB) (p70 ribosomal S6 kinase beta) (p70 S6Kbeta) (S6K2). | |||||