Please be patient as the page loads
|
GANAB_HUMAN
|
||||||
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GANAB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
|
GANAB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997308 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BHN3, O08794, Q80U81, Q9CS53 | Gene names | Ganab, G2an, Kiaa0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha) (Alpha glucosidase 2). | |||||
|
GANC_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.988183 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TET4, Q52LQ4, Q8IWZ0, Q8IZM4, Q8IZM5 | Gene names | GANC | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.20). | |||||
|
GANC_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.988083 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BVW0, Q8BH03 | Gene names | Ganc | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.-). | |||||
|
LYAG_HUMAN
|
||||||
| θ value | 9.37149e-90 (rank : 5) | NC score | 0.899420 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
LYAG_MOUSE
|
||||||
| θ value | 5.14231e-88 (rank : 6) | NC score | 0.899057 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
|
MGA_HUMAN
|
||||||
| θ value | 3.45727e-76 (rank : 7) | NC score | 0.864173 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
SUIS_HUMAN
|
||||||
| θ value | 1.89707e-74 (rank : 8) | NC score | 0.866503 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14410 | Gene names | SI | |||
|
Domain Architecture |

|
|||||
| Description | Sucrase-isomaltase, intestinal [Contains: Sucrase (EC 3.2.1.48); Isomaltase (EC 3.2.1.10)]. | |||||
|
AT132_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.015874 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.084103 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.013806 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CKLF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.027280 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAZ6, Q8N7E5 | Gene names | CMTM2, CKLFSF2 | |||
|
Domain Architecture |
|
|||||
| Description | CKLF-like MARVEL transmembrane domain-containing protein 2 (Chemokine- like factor superfamily member 2). | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.006854 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.019836 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
KI21B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.006699 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.012010 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.006232 (rank : 33) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CS029_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.013074 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUQ7, O75229, Q7LE08, Q9BTA6, Q9Y5A4 | Gene names | C19orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 (NY-REN-24 antigen). | |||||
|
TNR8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.014675 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
CAH9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.008993 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16790 | Gene names | CA9, G250, MN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN) (P54/58N) (Renal cell carcinoma-associated antigen G250) (RCC- associated antigen G250) (pMW1). | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.022135 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.010681 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.003763 (rank : 34) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
APBA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.006259 (rank : 32) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.002335 (rank : 36) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
SMBT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.006512 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHJ3, Q96C73, Q9Y4Q9 | Gene names | SFMBT1, RU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1 (Renal ubiquitous protein 1). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.008411 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.009259 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.009941 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
SIX4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.003737 (rank : 35) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIU6 | Gene names | SIX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX4 (Sine oculis homeobox homolog 4). | |||||
|
WDR65_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.006587 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MR6, Q5TAI0 | Gene names | WDR65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 65. | |||||
|
TFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.065190 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04155 | Gene names | TFF1, BCEI, PS2 | |||
|
Domain Architecture |
|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein) (HP1.A) (Breast cancer estrogen-inducible protein) (PNR-2). | |||||
|
TFF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.068529 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08423, Q3TVE6 | Gene names | Tff1, Bcei, Ps2 | |||
|
Domain Architecture |
|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein). | |||||
|
TFF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.054668 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03403, Q15854 | Gene names | TFF2, SML1 | |||
|
Domain Architecture |
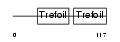
|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP) (Spasmolysin). | |||||
|
TFF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.067381 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07654 | Gene names | TFF3, ITF, TFI | |||
|
Domain Architecture |
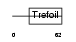
|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor) (hP1.B). | |||||
|
TFF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.083530 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62395, Q64352 | Gene names | Tff3, Itf | |||
|
Domain Architecture |
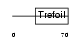
|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor). | |||||
|
GANAB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14697, Q8WTS9, Q9P0X0 | Gene names | GANAB, G2AN, KIAA0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha). | |||||
|
GANAB_MOUSE
|
||||||
| NC score | 0.997308 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BHN3, O08794, Q80U81, Q9CS53 | Gene names | Ganab, G2an, Kiaa0088 | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase AB precursor (EC 3.2.1.84) (Glucosidase II subunit alpha) (Alpha glucosidase 2). | |||||
|
GANC_HUMAN
|
||||||
| NC score | 0.988183 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TET4, Q52LQ4, Q8IWZ0, Q8IZM4, Q8IZM5 | Gene names | GANC | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.20). | |||||
|
GANC_MOUSE
|
||||||
| NC score | 0.988083 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BVW0, Q8BH03 | Gene names | Ganc | |||
|
Domain Architecture |

|
|||||
| Description | Neutral alpha-glucosidase C (EC 3.2.1.-). | |||||
|
LYAG_HUMAN
|
||||||
| NC score | 0.899420 (rank : 5) | θ value | 9.37149e-90 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10253, Q14351, Q16302 | Gene names | GAA | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase) (Aglucosidase alfa) [Contains: 76 kDa lysosomal alpha-glucosidase; 70 kDa lysosomal alpha-glucosidase]. | |||||
|
LYAG_MOUSE
|
||||||
| NC score | 0.899057 (rank : 6) | θ value | 5.14231e-88 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70699, Q3UJB2, Q8BGI6, Q91Z45 | Gene names | Gaa | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal alpha-glucosidase precursor (EC 3.2.1.20) (Acid maltase). | |||||
|
SUIS_HUMAN
|
||||||
| NC score | 0.866503 (rank : 7) | θ value | 1.89707e-74 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14410 | Gene names | SI | |||
|
Domain Architecture |

|
|||||
| Description | Sucrase-isomaltase, intestinal [Contains: Sucrase (EC 3.2.1.48); Isomaltase (EC 3.2.1.10)]. | |||||
|
MGA_HUMAN
|
||||||
| NC score | 0.864173 (rank : 8) | θ value | 3.45727e-76 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43451 | Gene names | MGAM, MGA, MGAML | |||
|
Domain Architecture |

|
|||||
| Description | Maltase-glucoamylase, intestinal [Includes: Maltase (EC 3.2.1.20) (Alpha-glucosidase); Glucoamylase (EC 3.2.1.3) (Glucan 1,4-alpha- glucosidase)]. | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.084103 (rank : 9) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
TFF3_MOUSE
|
||||||
| NC score | 0.083530 (rank : 10) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62395, Q64352 | Gene names | Tff3, Itf | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor). | |||||
|
TFF1_MOUSE
|
||||||
| NC score | 0.068529 (rank : 11) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08423, Q3TVE6 | Gene names | Tff1, Bcei, Ps2 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein). | |||||
|
TFF3_HUMAN
|
||||||
| NC score | 0.067381 (rank : 12) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07654 | Gene names | TFF3, ITF, TFI | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 3 precursor (Intestinal trefoil factor) (hP1.B). | |||||
|
TFF1_HUMAN
|
||||||
| NC score | 0.065190 (rank : 13) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04155 | Gene names | TFF1, BCEI, PS2 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 1 precursor (pS2 protein) (HP1.A) (Breast cancer estrogen-inducible protein) (PNR-2). | |||||
|
TFF2_HUMAN
|
||||||
| NC score | 0.054668 (rank : 14) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03403, Q15854 | Gene names | TFF2, SML1 | |||
|
Domain Architecture |

|
|||||
| Description | Trefoil factor 2 precursor (Spasmolytic polypeptide) (SP) (Spasmolysin). | |||||
|
CKLF2_HUMAN
|
||||||
| NC score | 0.027280 (rank : 15) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAZ6, Q8N7E5 | Gene names | CMTM2, CKLFSF2 | |||
|
Domain Architecture |

|
|||||
| Description | CKLF-like MARVEL transmembrane domain-containing protein 2 (Chemokine- like factor superfamily member 2). | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.022135 (rank : 16) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.019836 (rank : 17) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
AT132_HUMAN
|
||||||
| NC score | 0.015874 (rank : 18) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQ11, O75700, Q5JXY2 | Gene names | ATP13A2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
TNR8_MOUSE
|
||||||
| NC score | 0.014675 (rank : 19) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.013806 (rank : 20) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CS029_HUMAN
|
||||||
| NC score | 0.013074 (rank : 21) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUQ7, O75229, Q7LE08, Q9BTA6, Q9Y5A4 | Gene names | C19orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 (NY-REN-24 antigen). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.012010 (rank : 22) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.010681 (rank : 23) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.009941 (rank : 24) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.009259 (rank : 25) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CAH9_HUMAN
|
||||||
| NC score | 0.008993 (rank : 26) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16790 | Gene names | CA9, G250, MN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN) (P54/58N) (Renal cell carcinoma-associated antigen G250) (RCC- associated antigen G250) (pMW1). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.008411 (rank : 27) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.006854 (rank : 28) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
KI21B_MOUSE
|
||||||
| NC score | 0.006699 (rank : 29) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
WDR65_HUMAN
|
||||||
| NC score | 0.006587 (rank : 30) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MR6, Q5TAI0 | Gene names | WDR65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 65. | |||||
|
SMBT1_HUMAN
|
||||||
| NC score | 0.006512 (rank : 31) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHJ3, Q96C73, Q9Y4Q9 | Gene names | SFMBT1, RU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 1 (Renal ubiquitous protein 1). | |||||
|
APBA3_HUMAN
|
||||||
| NC score | 0.006259 (rank : 32) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.006232 (rank : 33) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.003763 (rank : 34) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
SIX4_HUMAN
|
||||||
| NC score | 0.003737 (rank : 35) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIU6 | Gene names | SIX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX4 (Sine oculis homeobox homolog 4). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.002335 (rank : 36) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||