Please be patient as the page loads
|
RYR2_HUMAN
|
||||||
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RYR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.960515 (rank : 3) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 175 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.975845 (rank : 2) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
ITPR1_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 4) | NC score | 0.551793 (rank : 4) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 3.61944e-33 (rank : 5) | NC score | 0.547645 (rank : 5) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR3_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 6) | NC score | 0.536082 (rank : 6) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR3_HUMAN
|
||||||
| θ value | 3.50572e-28 (rank : 7) | NC score | 0.532386 (rank : 7) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR2_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 8) | NC score | 0.530770 (rank : 8) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ITPR2_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 9) | NC score | 0.530373 (rank : 9) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
RN123_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 10) | NC score | 0.385322 (rank : 10) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5XPI4, Q5I022, Q6PFW4, Q71RH0, Q8IW18, Q9H0M8, Q9H5L8, Q9H9T2 | Gene names | RNF123, KPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
RN123_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 11) | NC score | 0.384725 (rank : 11) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5XPI3, Q6PGE0 | Gene names | Rnf123, Kpc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 12) | NC score | 0.134068 (rank : 18) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 13) | NC score | 0.131565 (rank : 19) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
ASH2L_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 14) | NC score | 0.268550 (rank : 12) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UBL3, O60659, O60660, Q96B62 | Gene names | ASH2L | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
ASH2L_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 15) | NC score | 0.267962 (rank : 13) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
RANB9_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 16) | NC score | 0.229152 (rank : 15) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
RANB9_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 17) | NC score | 0.242435 (rank : 14) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
HNRL1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 18) | NC score | 0.153746 (rank : 16) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 19) | NC score | 0.153470 (rank : 17) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.096737 (rank : 21) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 21) | NC score | 0.024030 (rank : 101) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 22) | NC score | 0.027944 (rank : 95) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
CABP7_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.096707 (rank : 22) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.096707 (rank : 23) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.100066 (rank : 20) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.092515 (rank : 25) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CALM_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.087480 (rank : 30) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.087480 (rank : 31) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
TRIM9_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.051346 (rank : 70) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9C026, Q92557, Q96D24, Q96NI4, Q9C025, Q9C027 | Gene names | TRIM9, KIAA0282, RNF91 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9 (RING finger protein 91). | |||||
|
ADA30_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.015382 (rank : 133) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.085742 (rank : 32) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
MLRV_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.090892 (rank : 27) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
TRIM9_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.051425 (rank : 69) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C7M3, Q6ZQE5, Q80WT6, Q8C7Z4, Q8CC32, Q8CEG2, Q99PQ3 | Gene names | Trim9, Kiaa0282 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.025141 (rank : 99) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.065157 (rank : 49) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.017276 (rank : 128) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.045687 (rank : 77) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MLR5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.090993 (rank : 26) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |
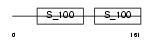
|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
CALL6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.079802 (rank : 38) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
RHBD4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.053511 (rank : 65) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P58872 | Gene names | RHBDL4, VRHO | |||
|
Domain Architecture |
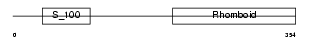
|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
CABP2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.088388 (rank : 28) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
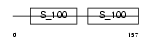
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
MLRS_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.081986 (rank : 36) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |
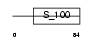
|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
RHBD4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.052322 (rank : 67) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |
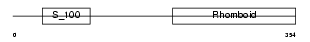
|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
SDF2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.051064 (rank : 71) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99470, Q9BQ79 | Gene names | SDF2 | |||
|
Domain Architecture |
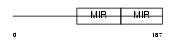
|
|||||
| Description | Stromal cell-derived factor 2 precursor (SDF-2). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.087669 (rank : 29) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
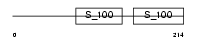
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CAN2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.021960 (rank : 106) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
PCAT1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.025103 (rank : 100) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NF37, Q1HAQ1, Q7Z4G6, Q8N3U7, Q8WUL8, Q9GZW6 | Gene names | AYTL2, LPCAT1, PFAAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (Acyltransferase-like 2) (Phosphonoformate immuno-associated protein 3). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.023286 (rank : 102) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
MEP50_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.017886 (rank : 123) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99J09, Q3TFJ1, Q8BSH8, Q9CZY5 | Gene names | Wdr77, Mep50 | |||
|
Domain Architecture |
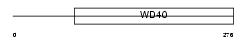
|
|||||
| Description | Methylosome protein 50 (MEP50 protein) (WD repeat protein 77). | |||||
|
MLRV_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.082663 (rank : 34) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |
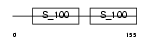
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.019121 (rank : 117) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
SDF2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.048861 (rank : 75) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCT5, P97307 | Gene names | Sdf2 | |||
|
Domain Architecture |
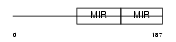
|
|||||
| Description | Stromal cell-derived factor 2 precursor (SDF-2). | |||||
|
SOMA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.041995 (rank : 79) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01241, Q14405, Q16631, Q5EB53, Q9HBZ1, Q9UMJ7, Q9UNL5 | Gene names | GH1 | |||
|
Domain Architecture |
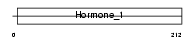
|
|||||
| Description | Somatotropin precursor (Growth hormone) (GH) (GH-N) (Pituitary growth hormone) (Growth hormone 1). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.016581 (rank : 130) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.022298 (rank : 104) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.028517 (rank : 94) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
CABP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.079602 (rank : 40) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
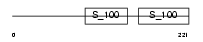
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CH60_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.021791 (rank : 107) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63038, P19226, P19227, P97602 | Gene names | Hspd1, Hsp60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (HSP-65). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.010609 (rank : 153) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
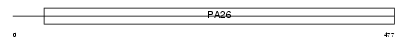
SESN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.017594 (rank : 127) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58004, Q96SI5 | Gene names | SESN2, SEST2 | |||
|
Domain Architecture |
|
|||||
| Description | Sestrin-2 (Hi95). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.013683 (rank : 139) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
SMRCD_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.009881 (rank : 158) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.031893 (rank : 91) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
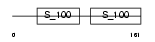
CABP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.078916 (rank : 43) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.085319 (rank : 33) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
LETM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.026238 (rank : 96) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.054857 (rank : 64) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.061785 (rank : 54) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
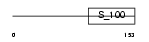
MLRA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.082496 (rank : 35) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MYL3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.055679 (rank : 61) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.021969 (rank : 105) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.017603 (rank : 126) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PLSL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.039336 (rank : 83) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
PLSL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.039428 (rank : 82) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
POGK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.010631 (rank : 152) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TC5, Q8BPJ3, Q8C887 | Gene names | Pogk, Kiaa1513 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with KRAB domain. | |||||
|
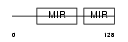
SDF2L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.043488 (rank : 78) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESP1, Q9ESP2 | Gene names | Sdf2l1 | |||
|
Domain Architecture |
|
|||||
| Description | Stromal cell-derived factor 2-like protein 1 precursor (SDF2-like protein 1). | |||||
|
ST17A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | -0.000939 (rank : 188) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UEE5, Q8IVC8 | Gene names | STK17A, DRAK1 | |||
|
Domain Architecture |
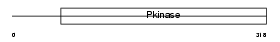
|
|||||
| Description | Serine/threonine-protein kinase 17A (EC 2.7.11.1) (DAP kinase-related apoptosis-inducing protein kinase 1). | |||||
|
TBCD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.017211 (rank : 129) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
CALN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.079467 (rank : 42) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
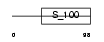
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.079511 (rank : 41) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
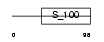
|
|||||
| Description | Calneuron-1. | |||||
|
CAN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.019671 (rank : 114) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.021781 (rank : 108) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.073451 (rank : 46) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
DOCK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.010551 (rank : 154) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14185 | Gene names | DOCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180). | |||||
|
IQGA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.014603 (rank : 137) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
MLRA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.080998 (rank : 37) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |
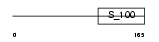
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MYL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.053387 (rank : 66) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P08590, Q9NRS8 | Gene names | MYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain) (Cardiac myosin light chain-1) (CMLC1). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.031151 (rank : 92) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
CD33_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.012149 (rank : 147) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q63994, Q63997 | Gene names | Cd33 | |||
|
Domain Architecture |
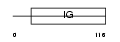
|
|||||
| Description | Myeloid cell surface antigen CD33 precursor (Siglec-3). | |||||
|
HMGN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.039154 (rank : 84) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05114, Q3KQR8 | Gene names | HMGN1, HMG14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nonhistone chromosomal protein HMG-14 (High-mobility group nucleosome- binding domain-containing protein 1). | |||||
|
PPE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.021011 (rank : 110) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14829, O15253, Q9NU21, Q9UJH0 | Gene names | PPEF1, PPEF, PPP7C | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 1 (EC 3.1.3.16) (PPEF-1) (Protein phosphatase with EF calcium-binding domain) (PPEF) (Serine/threonine-protein phosphatase 7) (PP7). | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.017735 (rank : 124) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.017676 (rank : 125) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.018054 (rank : 120) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
CAN3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.021460 (rank : 109) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
EFHB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.041430 (rank : 80) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
EGLX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.033756 (rank : 88) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NXG6, Q6PAG6, Q8TCJ9, Q8WV55, Q96F22, Q9BW77 | Gene names | PH4 | |||
|
Domain Architecture |
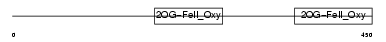
|
|||||
| Description | Putative HIF-prolyl hydroxylase PH-4 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl 4-hydroxylase). | |||||
|
EGLX_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.032144 (rank : 90) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BG58, Q8CAF1, Q9D499 | Gene names | Ph4 | |||
|
Domain Architecture |
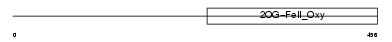
|
|||||
| Description | Putative HIF-prolyl hydroxylase PH-4 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl 4-hydroxylase). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.020946 (rank : 111) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.015193 (rank : 135) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.017905 (rank : 122) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
POT14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.012473 (rank : 146) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.003032 (rank : 183) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
AIF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.036336 (rank : 86) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55008, Q9UKS9 | Gene names | AIF1, G1, IBA1 | |||
|
Domain Architecture |
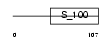
|
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1) (Protein G1). | |||||
|
CPNS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.023089 (rank : 103) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.006728 (rank : 172) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
LIMK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | -0.000714 (rank : 187) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53671, Q7KZ80, Q96E10, Q99464 | Gene names | LIMK2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain kinase 2 (EC 2.7.11.1) (LIMK-2). | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.019136 (rank : 116) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
ONCO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.050876 (rank : 72) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |
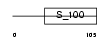
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.019298 (rank : 115) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
SDF2L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.037981 (rank : 85) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCN8, Q9BRI5 | Gene names | SDF2L1 | |||
|
Domain Architecture |
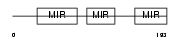
|
|||||
| Description | Stromal cell-derived factor 2-like protein 1 precursor (SDF2-like protein 1) (PWP1-interacting protein 8). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.006942 (rank : 169) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
UNC5B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.013662 (rank : 141) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
WDR7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.010365 (rank : 156) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4E6, Q86UX5, Q86VP2, Q96PS7 | Gene names | WDR7, KIAA0541, TRAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 7 (TGF-beta resistance-associated protein TRAG) (Rabconnectin-3 beta). | |||||
|
ASM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.011215 (rank : 149) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17405, P17406, Q13811, Q16837, Q16841 | Gene names | SMPD1, ASM | |||
|
Domain Architecture |
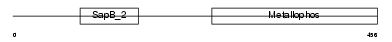
|
|||||
| Description | Sphingomyelin phosphodiesterase precursor (EC 3.1.4.12) (Acid sphingomyelinase) (aSMase). | |||||
|
CAYP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.035376 (rank : 87) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.007438 (rank : 168) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.029616 (rank : 93) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.005895 (rank : 178) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.006768 (rank : 171) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
N4BP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.017959 (rank : 121) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
POK7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.005199 (rank : 181) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QC07, Q6QAI9, Q96PI5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pol protein (HERV-K18 Pol protein) (HERV-K110 Pol protein) (HERV-K(C1a) Pol protein) [Includes: Reverse transcriptase (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H)]. | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.006841 (rank : 170) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.010951 (rank : 150) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.018307 (rank : 119) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.063876 (rank : 53) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
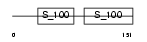
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
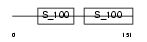
TNNC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.064151 (rank : 51) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.016267 (rank : 131) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
UNC5B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.013668 (rank : 140) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
ZN366_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | -0.002145 (rank : 190) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N895, Q7RTV4 | Gene names | ZNF366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 366. | |||||
|
ZN586_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.000266 (rank : 185) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 637 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NXT0 | Gene names | ZNF586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 586. | |||||
|
ANKR5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.009611 (rank : 159) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D2J7 | Gene names | Ankrd5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.006706 (rank : 173) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CAN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.019782 (rank : 112) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
CCD38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.012569 (rank : 145) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.009036 (rank : 161) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CND3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.045830 (rank : 76) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
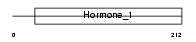
CSH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.015976 (rank : 132) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01243, Q14407 | Gene names | CSH1 | |||
|
Domain Architecture |
|
|||||
| Description | Chorionic somatomammotropin hormone precursor (Choriomammotropin) (Lactogen). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.012821 (rank : 144) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
SNW1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.011217 (rank : 148) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CSN1 | Gene names | Snw1, Skiip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski- interacting protein). | |||||
|
SPRY4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.040663 (rank : 81) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91WK1, Q8BUS6, Q9D651 | Gene names | Spryd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPRY domain-containing protein 4. | |||||
|
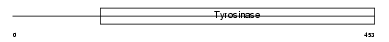
TYRO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.007958 (rank : 164) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11344 | Gene names | Tyr | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosinase precursor (EC 1.14.18.1) (Monophenol monooxygenase) (Albino locus protein). | |||||
|
ZFP59_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | -0.001708 (rank : 189) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16373, Q61849 | Gene names | Zfp59, Mfg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 59 (Zfp-59) (Zinc finger protein Mfg-2). | |||||
|
ARHG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.005859 (rank : 179) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NR81, Q6NUN3, Q7Z4U2, Q7Z5T2, Q9H7T4 | Gene names | ARHGEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3 (Exchange factor found in platelets and leukemic and neuronal tissues) (XPLN). | |||||
|
ARHG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.005856 (rank : 180) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91X46, Q8CDM0, Q91VY4, Q99K14, Q9DC31 | Gene names | Arhgef3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3. | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.007721 (rank : 166) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BSH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.000204 (rank : 186) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q810B3 | Gene names | Bsx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific homeobox protein homolog. | |||||
|
CALM4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.065467 (rank : 48) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CEP72_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.010483 (rank : 155) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P209, Q9BV03, Q9BWM3, Q9NVR4 | Gene names | CEP72, KIAA1519 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.014616 (rank : 136) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.006412 (rank : 174) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
DPOD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.015336 (rank : 134) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWP8 | Gene names | Pold4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase subunit delta 4 (DNA polymerase subunit delta p12). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.008361 (rank : 163) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
EXOC8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.009178 (rank : 160) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYI6, Q5TE82 | Gene names | EXOC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 8 (Exocyst complex 84 kDa subunit). | |||||
|
FAK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | -0.002272 (rank : 192) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.003896 (rank : 182) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
LETM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.019695 (rank : 113) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z2I0, Q5PQC5, Q7TMK8, Q80ZX6, Q8CGJ3, Q8K5E5 | Gene names | Letm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
LYST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.008578 (rank : 162) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.013324 (rank : 142) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
MLE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.049632 (rank : 74) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.006373 (rank : 175) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.013284 (rank : 143) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.019057 (rank : 118) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.010924 (rank : 151) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.001490 (rank : 184) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
RM40_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.033712 (rank : 89) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2Q5, Q9CS52 | Gene names | Mrpl40, Nlvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L40, mitochondrial precursor (L40mt) (MRP-40) (Nuclear localization signal-containing protein deleted in velocardiofacial syndrome homolog). | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.006222 (rank : 177) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.014386 (rank : 138) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.007803 (rank : 165) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
UACA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.006271 (rank : 176) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
UBCP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.026229 (rank : 97) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVY7, Q96DK5 | Gene names | UBLCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
UBCP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.026226 (rank : 98) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGR9, Q52L62, Q5XJH1, Q6PH96, Q99LT3 | Gene names | Ublcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
UT14B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.007592 (rank : 167) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
VATC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.010151 (rank : 157) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1G3 | Gene names | Atp6v1c1, Atp6c, Vatc | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase subunit C (EC 3.6.3.14) (V-ATPase C subunit) (Vacuolar proton pump C subunit). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | -0.002221 (rank : 191) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
CABP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.064902 (rank : 50) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.063878 (rank : 52) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CETN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.057124 (rank : 57) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
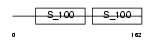
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.056760 (rank : 58) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.057599 (rank : 55) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.057569 (rank : 56) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.055050 (rank : 62) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.054921 (rank : 63) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
EFCB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.070780 (rank : 47) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
HNRPU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.095155 (rank : 24) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
MLRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.077341 (rank : 44) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |
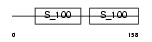
|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.076807 (rank : 45) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |
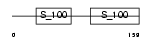
|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.079729 (rank : 39) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MYL6B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.051481 (rank : 68) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
SPRY3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.050027 (rank : 73) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NCJ5, Q96SK5 | Gene names | SPRYD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPRY domain-containing protein 3. | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.056382 (rank : 60) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.056547 (rank : 59) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
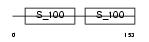
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 175 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.975845 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.960515 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.551793 (rank : 4) | θ value | 2.50655e-34 (rank : 4) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.547645 (rank : 5) | θ value | 3.61944e-33 (rank : 5) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR3_MOUSE
|
||||||
| NC score | 0.536082 (rank : 6) | θ value | 4.14116e-29 (rank : 6) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR3_HUMAN
|
||||||
| NC score | 0.532386 (rank : 7) | θ value | 3.50572e-28 (rank : 7) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
ITPR2_MOUSE
|
||||||
| NC score | 0.530770 (rank : 8) | θ value | 8.63488e-27 (rank : 8) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ITPR2_HUMAN
|
||||||
| NC score | 0.530373 (rank : 9) | θ value | 1.12775e-26 (rank : 9) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
RN123_HUMAN
|
||||||
| NC score | 0.385322 (rank : 10) | θ value | 1.16975e-15 (rank : 10) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5XPI4, Q5I022, Q6PFW4, Q71RH0, Q8IW18, Q9H0M8, Q9H5L8, Q9H9T2 | Gene names | RNF123, KPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
RN123_MOUSE
|
||||||
| NC score | 0.384725 (rank : 11) | θ value | 1.16975e-15 (rank : 11) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5XPI3, Q6PGE0 | Gene names | Rnf123, Kpc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
ASH2L_HUMAN
|
||||||
| NC score | 0.268550 (rank : 12) | θ value | 1.09739e-05 (rank : 14) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UBL3, O60659, O60660, Q96B62 | Gene names | ASH2L | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
ASH2L_MOUSE
|
||||||
| NC score | 0.267962 (rank : 13) | θ value | 1.09739e-05 (rank : 15) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91X20, Q3UIF9, Q9Z2X4 | Gene names | Ash2l | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
RANB9_MOUSE
|
||||||
| NC score | 0.242435 (rank : 14) | θ value | 0.000158464 (rank : 17) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
RANB9_HUMAN
|
||||||
| NC score | 0.229152 (rank : 15) | θ value | 0.000158464 (rank : 16) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
HNRL1_HUMAN
|
||||||
| NC score | 0.153746 (rank : 16) | θ value | 0.000786445 (rank : 18) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.153470 (rank : 17) | θ value | 0.000786445 (rank : 19) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.134068 (rank : 18) | θ value | 1.53129e-07 (rank : 12) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.131565 (rank : 19) | θ value | 2.61198e-07 (rank : 13) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.100066 (rank : 20) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.096737 (rank : 21) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP7_HUMAN
|
||||||
| NC score | 0.096707 (rank : 22) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| NC score | 0.096707 (rank : 23) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
HNRPU_HUMAN
|
||||||
| NC score | 0.095155 (rank : 24) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.092515 (rank : 25) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
MLR5_HUMAN
|
||||||
| NC score | 0.090993 (rank : 26) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |

|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
MLRV_HUMAN
|
||||||
| NC score | 0.090892 (rank : 27) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.088388 (rank : 28) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.087669 (rank : 29) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
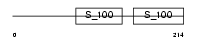
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.087480 (rank : 30) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.087480 (rank : 31) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.085742 (rank : 32) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.085319 (rank : 33) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
MLRV_MOUSE
|
||||||
| NC score | 0.082663 (rank : 34) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRA_MOUSE
|
||||||
| NC score | 0.082496 (rank : 35) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRS_MOUSE
|
||||||
| NC score | 0.081986 (rank : 36) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
MLRA_HUMAN
|
||||||
| NC score | 0.080998 (rank : 37) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
CALL6_HUMAN
|
||||||
| NC score | 0.079802 (rank : 38) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
MLRN_MOUSE
|
||||||
| NC score | 0.079729 (rank : 39) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.079602 (rank : 40) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.079511 (rank : 41) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.079467 (rank : 42) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.078916 (rank : 43) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
MLRM_HUMAN
|
||||||
| NC score | 0.077341 (rank : 44) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| NC score | 0.076807 (rank : 45) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.073451 (rank : 46) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
EFCB2_MOUSE
|
||||||
| NC score | 0.070780 (rank : 47) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.065467 (rank : 48) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.065157 (rank : 49) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.064902 (rank : 50) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.064151 (rank : 51) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.063878 (rank : 52) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.063876 (rank : 53) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.061785 (rank : 54) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.057599 (rank : 55) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.057569 (rank : 56) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.057124 (rank : 57) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.056760 (rank : 58) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.056547 (rank : 59) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.056382 (rank : 60) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
MYL3_MOUSE
|
||||||
| NC score | 0.055679 (rank : 61) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.055050 (rank : 62) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| NC score | 0.054921 (rank : 63) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.054857 (rank : 64) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
RHBD4_HUMAN
|
||||||
| NC score | 0.053511 (rank : 65) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P58872 | Gene names | RHBDL4, VRHO | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
MYL3_HUMAN
|
||||||
| NC score | 0.053387 (rank : 66) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P08590, Q9NRS8 | Gene names | MYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain) (Cardiac myosin light chain-1) (CMLC1). | |||||
|
RHBD4_MOUSE
|
||||||
| NC score | 0.052322 (rank : 67) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P58873 | Gene names | Rhbdl4, Vrho | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 4 (EC 3.4.21.105) (Ventrhoid transmembrane protein). | |||||
|
MYL6B_HUMAN
|
||||||
| NC score | 0.051481 (rank : 68) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
TRIM9_MOUSE
|
||||||
| NC score | 0.051425 (rank : 69) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C7M3, Q6ZQE5, Q80WT6, Q8C7Z4, Q8CC32, Q8CEG2, Q99PQ3 | Gene names | Trim9, Kiaa0282 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9. | |||||
|
TRIM9_HUMAN
|
||||||
| NC score | 0.051346 (rank : 70) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9C026, Q92557, Q96D24, Q96NI4, Q9C025, Q9C027 | Gene names | TRIM9, KIAA0282, RNF91 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 9 (RING finger protein 91). | |||||
|
SDF2_HUMAN
|
||||||
| NC score | 0.051064 (rank : 71) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99470, Q9BQ79 | Gene names | SDF2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromal cell-derived factor 2 precursor (SDF-2). | |||||
|
ONCO_HUMAN
|
||||||
| NC score | 0.050876 (rank : 72) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
SPRY3_HUMAN
|
||||||
| NC score | 0.050027 (rank : 73) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NCJ5, Q96SK5 | Gene names | SPRYD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPRY domain-containing protein 3. | |||||
|
MLE1_HUMAN
|
||||||
| NC score | 0.049632 (rank : 74) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
SDF2_MOUSE
|
||||||
| NC score | 0.048861 (rank : 75) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DCT5, P97307 | Gene names | Sdf2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromal cell-derived factor 2 precursor (SDF-2). | |||||
|
CND3_HUMAN
|
||||||
| NC score | 0.045830 (rank : 76) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.045687 (rank : 77) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SDF2L_MOUSE
|
||||||
| NC score | 0.043488 (rank : 78) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESP1, Q9ESP2 | Gene names | Sdf2l1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromal cell-derived factor 2-like protein 1 precursor (SDF2-like protein 1). | |||||
|
SOMA_HUMAN
|
||||||
| NC score | 0.041995 (rank : 79) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01241, Q14405, Q16631, Q5EB53, Q9HBZ1, Q9UMJ7, Q9UNL5 | Gene names | GH1 | |||
|
Domain Architecture |

|
|||||
| Description | Somatotropin precursor (Growth hormone) (GH) (GH-N) (Pituitary growth hormone) (Growth hormone 1). | |||||
|
EFHB_MOUSE
|
||||||
| NC score | 0.041430 (rank : 80) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
SPRY4_MOUSE
|
||||||
| NC score | 0.040663 (rank : 81) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91WK1, Q8BUS6, Q9D651 | Gene names | Spryd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPRY domain-containing protein 4. | |||||
|
PLSL_MOUSE
|
||||||
| NC score | 0.039428 (rank : 82) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PLSL_HUMAN
|
||||||
| NC score | 0.039336 (rank : 83) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
HMGN1_HUMAN
|
||||||
| NC score | 0.039154 (rank : 84) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05114, Q3KQR8 | Gene names | HMGN1, HMG14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nonhistone chromosomal protein HMG-14 (High-mobility group nucleosome- binding domain-containing protein 1). | |||||
|
SDF2L_HUMAN
|
||||||
| NC score | 0.037981 (rank : 85) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCN8, Q9BRI5 | Gene names | SDF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromal cell-derived factor 2-like protein 1 precursor (SDF2-like protein 1) (PWP1-interacting protein 8). | |||||
|
AIF1_HUMAN
|
||||||
| NC score | 0.036336 (rank : 86) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55008, Q9UKS9 | Gene names | AIF1, G1, IBA1 | |||
|
Domain Architecture |

|
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1) (Protein G1). | |||||
|
CAYP2_HUMAN
|
||||||
| NC score | 0.035376 (rank : 87) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
EGLX_HUMAN
|
||||||
| NC score | 0.033756 (rank : 88) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NXG6, Q6PAG6, Q8TCJ9, Q8WV55, Q96F22, Q9BW77 | Gene names | PH4 | |||
|
Domain Architecture |
|
|||||
| Description | Putative HIF-prolyl hydroxylase PH-4 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl 4-hydroxylase). | |||||
|
RM40_MOUSE
|
||||||
| NC score | 0.033712 (rank : 89) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2Q5, Q9CS52 | Gene names | Mrpl40, Nlvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L40, mitochondrial precursor (L40mt) (MRP-40) (Nuclear localization signal-containing protein deleted in velocardiofacial syndrome homolog). | |||||
|
EGLX_MOUSE
|
||||||
| NC score | 0.032144 (rank : 90) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BG58, Q8CAF1, Q9D499 | Gene names | Ph4 | |||
|
Domain Architecture |

|
|||||
| Description | Putative HIF-prolyl hydroxylase PH-4 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl 4-hydroxylase). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.031893 (rank : 91) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.031151 (rank : 92) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.029616 (rank : 93) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.028517 (rank : 94) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.027944 (rank : 95) | θ value | 0.0193708 (rank : 22) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
LETM1_HUMAN
|
||||||
| NC score | 0.026238 (rank : 96) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
UBCP1_HUMAN
|
||||||
| NC score | 0.026229 (rank : 97) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVY7, Q96DK5 | Gene names | UBLCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
UBCP1_MOUSE
|
||||||
| NC score | 0.026226 (rank : 98) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGR9, Q52L62, Q5XJH1, Q6PH96, Q99LT3 | Gene names | Ublcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like domain-containing CTD phosphatase 1 (EC 3.1.3.16). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.025141 (rank : 99) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
PCAT1_HUMAN
|
||||||
| NC score | 0.025103 (rank : 100) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NF37, Q1HAQ1, Q7Z4G6, Q8N3U7, Q8WUL8, Q9GZW6 | Gene names | AYTL2, LPCAT1, PFAAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (Acyltransferase-like 2) (Phosphonoformate immuno-associated protein 3). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.024030 (rank : 101) | θ value | 0.0193708 (rank : 21) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.023286 (rank : 102) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
CPNS2_MOUSE
|
||||||
| NC score | 0.023089 (rank : 103) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D7J7 | Gene names | Capns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.022298 (rank : 104) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.021969 (rank : 105) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
CAN2_MOUSE
|
||||||
| NC score | 0.021960 (rank : 106) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
CH60_MOUSE
|
||||||
| NC score | 0.021791 (rank : 107) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63038, P19226, P19227, P97602 | Gene names | Hspd1, Hsp60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60 kDa heat shock protein, mitochondrial precursor (Hsp60) (60 kDa chaperonin) (CPN60) (Heat shock protein 60) (HSP-60) (Mitochondrial matrix protein P1) (HSP-65). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.021781 (rank : 108) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
CAN3_MOUSE
|
||||||
| NC score | 0.021460 (rank : 109) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64691, Q9WUC5 | Gene names | Capn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3). | |||||
|
PPE1_HUMAN
|
||||||
| NC score | 0.021011 (rank : 110) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14829, O15253, Q9NU21, Q9UJH0 | Gene names | PPEF1, PPEF, PPP7C | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 1 (EC 3.1.3.16) (PPEF-1) (Protein phosphatase with EF calcium-binding domain) (PPEF) (Serine/threonine-protein phosphatase 7) (PP7). | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.020946 (rank : 111) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
CAN3_HUMAN
|
||||||
| NC score | 0.019782 (rank : 112) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P20807, Q9BTU4, Q9Y5S6, Q9Y5S7 | Gene names | CAPN3, CANP3, CANPL3, NCL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-3 (EC 3.4.22.-) (Calpain L3) (Calpain p94) (Calcium-activated neutral proteinase 3) (CANP 3) (Muscle-specific calcium-activated neutral protease 3) (nCL-1). | |||||
|
LETM1_MOUSE
|
||||||
| NC score | 0.019695 (rank : 113) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z2I0, Q5PQC5, Q7TMK8, Q80ZX6, Q8CGJ3, Q8K5E5 | Gene names | Letm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
CAN2_HUMAN
|
||||||
| NC score | 0.019671 (rank : 114) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.019298 (rank : 115) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.019136 (rank : 116) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.019121 (rank : 117) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.019057 (rank : 118) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.018307 (rank : 119) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.018054 (rank : 120) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
N4BP2_HUMAN
|
||||||
| NC score | 0.017959 (rank : 121) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.017905 (rank : 122) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
MEP50_MOUSE
|
||||||
| NC score | 0.017886 (rank : 123) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99J09, Q3TFJ1, Q8BSH8, Q9CZY5 | Gene names | Wdr77, Mep50 | |||
|
Domain Architecture |

|
|||||
| Description | Methylosome protein 50 (MEP50 protein) (WD repeat protein 77). | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.017735 (rank : 124) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.017676 (rank : 125) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.017603 (rank : 126) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
SESN2_HUMAN
|
||||||
| NC score | 0.017594 (rank : 127) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58004, Q96SI5 | Gene names | SESN2, SEST2 | |||
|
Domain Architecture |

|
|||||
| Description | Sestrin-2 (Hi95). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.017276 (rank : 128) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TBCD1_MOUSE
|
||||||
| NC score | 0.017211 (rank : 129) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.016581 (rank : 130) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.016267 (rank : 131) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
CSH_HUMAN
|
||||||
| NC score | 0.015976 (rank : 132) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01243, Q14407 | Gene names | CSH1 | |||
|
Domain Architecture |

|
|||||
| Description | Chorionic somatomammotropin hormone precursor (Choriomammotropin) (Lactogen). | |||||
|
ADA30_HUMAN
|
||||||
| NC score | 0.015382 (rank : 133) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
DPOD4_MOUSE
|
||||||
| NC score | 0.015336 (rank : 134) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWP8 | Gene names | Pold4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase subunit delta 4 (DNA polymerase subunit delta p12). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.015193 (rank : 135) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.014616 (rank : 136) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
IQGA1_MOUSE
|
||||||
| NC score | 0.014603 (rank : 137) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JKF1 | Gene names | Iqgap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP1. | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.014386 (rank : 138) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.013683 (rank : 139) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
UNC5B_MOUSE
|
||||||
| NC score | 0.013668 (rank : 140) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
UNC5B_HUMAN
|
||||||
| NC score | 0.013662 (rank : 141) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.013324 (rank : 142) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.013284 (rank : 143) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.012821 (rank : 144) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
CCD38_HUMAN
|
||||||
| NC score | 0.012569 (rank : 145) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
POT14_HUMAN
|
||||||
| NC score | 0.012473 (rank : 146) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
CD33_MOUSE
|
||||||
| NC score | 0.012149 (rank : 147) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q63994, Q63997 | Gene names | Cd33 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid cell surface antigen CD33 precursor (Siglec-3). | |||||
|
SNW1_MOUSE
|
||||||
| NC score | 0.011217 (rank : 148) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CSN1 | Gene names | Snw1, Skiip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNW domain-containing protein 1 (Nuclear protein SkiP) (Ski- interacting protein). | |||||
|
ASM_HUMAN
|
||||||
| NC score | 0.011215 (rank : 149) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17405, P17406, Q13811, Q16837, Q16841 | Gene names | SMPD1, ASM | |||
|
Domain Architecture |

|
|||||
| Description | Sphingomyelin phosphodiesterase precursor (EC 3.1.4.12) (Acid sphingomyelinase) (aSMase). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.010951 (rank : 150) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.010924 (rank : 151) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
POGK_MOUSE
|
||||||
| NC score | 0.010631 (rank : 152) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TC5, Q8BPJ3, Q8C887 | Gene names | Pogk, Kiaa1513 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with KRAB domain. | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.010609 (rank : 153) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DOCK1_HUMAN
|
||||||
| NC score | 0.010551 (rank : 154) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14185 | Gene names | DOCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 1 (180 kDa protein downstream of CRK) (DOCK180). | |||||
|
CEP72_HUMAN
|
||||||
| NC score | 0.010483 (rank : 155) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P209, Q9BV03, Q9BWM3, Q9NVR4 | Gene names | CEP72, KIAA1519 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
WDR7_HUMAN
|
||||||
| NC score | 0.010365 (rank : 156) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4E6, Q86UX5, Q86VP2, Q96PS7 | Gene names | WDR7, KIAA0541, TRAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 7 (TGF-beta resistance-associated protein TRAG) (Rabconnectin-3 beta). | |||||
|
VATC_MOUSE
|
||||||
| NC score | 0.010151 (rank : 157) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1G3 | Gene names | Atp6v1c1, Atp6c, Vatc | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase subunit C (EC 3.6.3.14) (V-ATPase C subunit) (Vacuolar proton pump C subunit). | |||||
|
SMRCD_MOUSE
|
||||||
| NC score | 0.009881 (rank : 158) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
ANKR5_MOUSE
|
||||||
| NC score | 0.009611 (rank : 159) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D2J7 | Gene names | Ankrd5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
EXOC8_HUMAN
|
||||||
| NC score | 0.009178 (rank : 160) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYI6, Q5TE82 | Gene names | EXOC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 8 (Exocyst complex 84 kDa subunit). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.009036 (rank : 161) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.008578 (rank : 162) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.008361 (rank : 163) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
TYRO_MOUSE
|
||||||
| NC score | 0.007958 (rank : 164) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11344 | Gene names | Tyr | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosinase precursor (EC 1.14.18.1) (Monophenol monooxygenase) (Albino locus protein). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.007803 (rank : 165) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.007721 (rank : 166) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
UT14B_MOUSE
|
||||||
| NC score | 0.007592 (rank : 167) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.007438 (rank : 168) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.006942 (rank : 169) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.006841 (rank : 170) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.006768 (rank : 171) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.006728 (rank : 172) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.006706 (rank : 173) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.006412 (rank : 174) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.006373 (rank : 175) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.006271 (rank : 176) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.006222 (rank : 177) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.005895 (rank : 178) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
ARHG3_HUMAN
|
||||||
| NC score | 0.005859 (rank : 179) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NR81, Q6NUN3, Q7Z4U2, Q7Z5T2, Q9H7T4 | Gene names | ARHGEF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3 (Exchange factor found in platelets and leukemic and neuronal tissues) (XPLN). | |||||
|
ARHG3_MOUSE
|
||||||
| NC score | 0.005856 (rank : 180) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91X46, Q8CDM0, Q91VY4, Q99K14, Q9DC31 | Gene names | Arhgef3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 3. | |||||
|
POK7_HUMAN
|
||||||
| NC score | 0.005199 (rank : 181) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QC07, Q6QAI9, Q96PI5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pol protein (HERV-K18 Pol protein) (HERV-K110 Pol protein) (HERV-K(C1a) Pol protein) [Includes: Reverse transcriptase (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H)]. | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.003896 (rank : 182) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.003032 (rank : 183) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.001490 (rank : 184) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
ZN586_HUMAN
|
||||||
| NC score | 0.000266 (rank : 185) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 637 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NXT0 | Gene names | ZNF586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 586. | |||||
|
BSH_MOUSE
|
||||||
| NC score | 0.000204 (rank : 186) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q810B3 | Gene names | Bsx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific homeobox protein homolog. | |||||
|
LIMK2_HUMAN
|
||||||
| NC score | -0.000714 (rank : 187) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53671, Q7KZ80, Q96E10, Q99464 | Gene names | LIMK2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain kinase 2 (EC 2.7.11.1) (LIMK-2). | |||||
|
ST17A_HUMAN
|
||||||
| NC score | -0.000939 (rank : 188) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UEE5, Q8IVC8 | Gene names | STK17A, DRAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 17A (EC 2.7.11.1) (DAP kinase-related apoptosis-inducing protein kinase 1). | |||||
|
ZFP59_MOUSE
|
||||||
| NC score | -0.001708 (rank : 189) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16373, Q61849 | Gene names | Zfp59, Mfg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 59 (Zfp-59) (Zinc finger protein Mfg-2). | |||||
|
ZN366_HUMAN
|
||||||
| NC score | -0.002145 (rank : 190) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N895, Q7RTV4 | Gene names | ZNF366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 366. | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | -0.002221 (rank : 191) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
FAK2_MOUSE
|
||||||
| NC score | -0.002272 (rank : 192) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 175 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||