Please be patient as the page loads
|
UBP47_HUMAN
|
||||||
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP47_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 149 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995727 (rank : 2) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 1.41672e-45 (rank : 3) | NC score | 0.876781 (rank : 3) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 4) | NC score | 0.861892 (rank : 4) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 5) | NC score | 0.839317 (rank : 6) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 6) | NC score | 0.856857 (rank : 5) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 6.82597e-32 (rank : 7) | NC score | 0.823503 (rank : 9) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 8) | NC score | 0.832109 (rank : 7) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 3.3877e-31 (rank : 9) | NC score | 0.763105 (rank : 16) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 10) | NC score | 0.753572 (rank : 19) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 11) | NC score | 0.771259 (rank : 13) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 12) | NC score | 0.754386 (rank : 18) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 1.2049e-28 (rank : 13) | NC score | 0.771126 (rank : 14) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 14) | NC score | 0.828521 (rank : 8) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 15) | NC score | 0.790106 (rank : 10) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 16) | NC score | 0.758303 (rank : 17) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 17) | NC score | 0.787479 (rank : 11) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 18) | NC score | 0.787479 (rank : 12) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 19) | NC score | 0.718962 (rank : 24) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 20) | NC score | 0.731624 (rank : 21) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 21) | NC score | 0.741748 (rank : 20) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 22) | NC score | 0.730253 (rank : 22) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 23) | NC score | 0.729509 (rank : 23) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 24) | NC score | 0.763255 (rank : 15) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 25) | NC score | 0.669239 (rank : 38) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 26) | NC score | 0.704101 (rank : 32) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 27) | NC score | 0.712767 (rank : 25) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 28) | NC score | 0.711444 (rank : 27) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 29) | NC score | 0.704580 (rank : 30) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 30) | NC score | 0.652918 (rank : 41) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 31) | NC score | 0.702761 (rank : 33) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 32) | NC score | 0.646367 (rank : 42) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 33) | NC score | 0.706532 (rank : 29) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 34) | NC score | 0.711258 (rank : 28) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 35) | NC score | 0.704145 (rank : 31) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 36) | NC score | 0.711655 (rank : 26) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 37) | NC score | 0.681379 (rank : 36) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 38) | NC score | 0.500159 (rank : 61) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 39) | NC score | 0.683049 (rank : 35) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 40) | NC score | 0.497089 (rank : 62) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 41) | NC score | 0.674140 (rank : 37) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 42) | NC score | 0.666238 (rank : 40) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 43) | NC score | 0.666946 (rank : 39) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 44) | NC score | 0.626355 (rank : 45) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 45) | NC score | 0.599430 (rank : 46) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 46) | NC score | 0.685327 (rank : 34) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 47) | NC score | 0.494888 (rank : 63) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 48) | NC score | 0.508630 (rank : 59) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 49) | NC score | 0.565076 (rank : 56) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP25_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 50) | NC score | 0.491565 (rank : 64) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 51) | NC score | 0.505165 (rank : 60) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 52) | NC score | 0.593485 (rank : 49) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 53) | NC score | 0.593511 (rank : 48) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 54) | NC score | 0.580835 (rank : 53) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 55) | NC score | 0.594966 (rank : 47) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 56) | NC score | 0.635695 (rank : 43) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 57) | NC score | 0.589904 (rank : 51) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 58) | NC score | 0.590076 (rank : 50) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 59) | NC score | 0.491486 (rank : 65) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 60) | NC score | 0.581687 (rank : 52) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 61) | NC score | 0.476370 (rank : 67) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 62) | NC score | 0.484494 (rank : 66) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 63) | NC score | 0.459603 (rank : 69) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 64) | NC score | 0.540227 (rank : 57) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 65) | NC score | 0.469074 (rank : 68) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 66) | NC score | 0.398724 (rank : 72) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 67) | NC score | 0.575642 (rank : 55) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 68) | NC score | 0.627468 (rank : 44) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 69) | NC score | 0.576747 (rank : 54) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 70) | NC score | 0.538127 (rank : 58) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
ASXL1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 71) | NC score | 0.081682 (rank : 74) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 72) | NC score | 0.080225 (rank : 75) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 73) | NC score | 0.035620 (rank : 92) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
SAFB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 74) | NC score | 0.037302 (rank : 90) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 75) | NC score | 0.436038 (rank : 70) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 0.125558 (rank : 76) | NC score | 0.027082 (rank : 97) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
SPT6H_HUMAN
|
||||||
| θ value | 0.163984 (rank : 77) | NC score | 0.063136 (rank : 78) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
SPT6H_MOUSE
|
||||||
| θ value | 0.163984 (rank : 78) | NC score | 0.062651 (rank : 79) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 79) | NC score | 0.043821 (rank : 87) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DLG4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 80) | NC score | 0.018125 (rank : 108) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 81) | NC score | 0.018164 (rank : 107) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
TREX2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 82) | NC score | 0.043498 (rank : 88) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQ50, Q45F08, Q9UN77 | Gene names | TREX2 | |||
|
Domain Architecture |
|
|||||
| Description | Three prime repair exonuclease 2 (EC 3.1.11.2) (3'-5' exonuclease TREX2). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 83) | NC score | 0.043231 (rank : 89) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 84) | NC score | 0.010481 (rank : 122) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 85) | NC score | 0.022298 (rank : 101) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 86) | NC score | 0.028810 (rank : 94) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
DLG1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 87) | NC score | 0.017288 (rank : 111) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.018104 (rank : 109) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
MGR4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.011176 (rank : 120) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 1.06291 (rank : 90) | NC score | 0.013392 (rank : 118) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
DLG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.016874 (rank : 112) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.048053 (rank : 85) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.021950 (rank : 104) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
SQSTM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.027977 (rank : 95) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
TOP3A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 95) | NC score | 0.016057 (rank : 114) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.062432 (rank : 80) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
GAGC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.022004 (rank : 103) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60829, Q6IBI1 | Gene names | PAGE4, GAGEC1 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen family C member 1 (Prostate-associated gene 4 protein) (PAGE-4) (PAGE-1) (GAGE-9). | |||||
|
STON1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.022233 (rank : 102) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
ZN583_HUMAN
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | -0.003417 (rank : 151) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96ND8, O14850, Q2NKK3 | Gene names | ZNF583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583 (Zinc finger protein L3-5). | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.010126 (rank : 125) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
FBX30_HUMAN
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.023488 (rank : 99) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TB52, Q9BXZ7 | Gene names | FBXO30, FBX30 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 30. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.036054 (rank : 91) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
N4BP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.022451 (rank : 100) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
NEK8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | -0.003106 (rank : 150) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZR4, Q3U498, Q9D685 | Gene names | Nek8, Jck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8). | |||||
|
TREX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.027687 (rank : 96) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1A9 | Gene names | Trex2 | |||
|
Domain Architecture |
|
|||||
| Description | Three prime repair exonuclease 2 (EC 3.1.11.2) (3'-5' exonuclease TREX2). | |||||
|
DBF4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.020898 (rank : 106) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ41, Q3TQP4, Q3V1K2, Q6NSQ1, Q9CXF2 | Gene names | Dbf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (MuDBF4). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.043906 (rank : 86) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
K1H2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.002851 (rank : 139) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.000081 (rank : 146) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NEK8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | -0.003436 (rank : 152) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SG6, Q2M1S6, Q8NDH1 | Gene names | NEK8, JCK, NEK12A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8) (NIMA-related kinase 12a). | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.007521 (rank : 127) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
ZN518_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.000073 (rank : 147) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
DEK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.034728 (rank : 93) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.049679 (rank : 84) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MYEF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.010160 (rank : 124) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2K5, Q6NUM5, Q7L388, Q7Z4B7, Q9H922, Q9NUQ1 | Gene names | MYEF2, KIAA1341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MST156). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.001745 (rank : 143) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
ATL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.003339 (rank : 135) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSK7, Q3U0C8 | Gene names | Adamtsl2, Kiaa0605, Tcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-like protein 2 precursor (ADAMTSL-2) (TSP1-repeats-containing protein 1) (TCP-1). | |||||
|
BNIP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.016699 (rank : 113) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54940 | Gene names | Bnip2, Nip2l | |||
|
Domain Architecture |
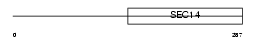
|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.003337 (rank : 136) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CT172_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.024987 (rank : 98) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CYC5, Q9CYF2, Q9DA26 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf172 homolog. | |||||
|
K1H2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.002010 (rank : 142) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |
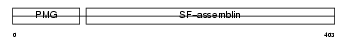
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
KV2A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.003347 (rank : 134) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01626 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region MOPC 167. | |||||
|
KV2E_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.005197 (rank : 132) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P03976 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region 17S29.1. | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.012828 (rank : 119) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
TRAF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.005462 (rank : 131) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00463 | Gene names | TRAF5, RNF84 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5 (RING finger protein 84). | |||||
|
ZN431_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | -0.004100 (rank : 153) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TF32, Q8IWC4 | Gene names | ZNF431, KIAA1969 | |||
|
Domain Architecture |
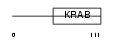
|
|||||
| Description | Zinc finger protein 431. | |||||
|
ZPI_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.002866 (rank : 138) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R121, Q8VCV8 | Gene names | Serpina10, Zpi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Z-dependent protease inhibitor precursor (PZ-dependent protease inhibitor) (PZI) (Serpin A10). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.010200 (rank : 123) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.002108 (rank : 141) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
GSTT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.015438 (rank : 115) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61133, Q61134, Q64472 | Gene names | Gstt2 | |||
|
Domain Architecture |
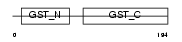
|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
K1H1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.002127 (rank : 140) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61765 | Gene names | Krtha1, Hka1, Krt1-1 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha1 (Hair keratin, type I Ha1) (HKA-1). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.021464 (rank : 105) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.017329 (rank : 110) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
SGCG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.013537 (rank : 117) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13326 | Gene names | SGCG | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-sarcoglycan (Gamma-SG) (35 kDa dystrophin-associated glycoprotein) (35DAG). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.010701 (rank : 121) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.000009 (rank : 148) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.001089 (rank : 145) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | -0.001472 (rank : 149) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GBA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.004826 (rank : 133) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H227, Q3MIH4, Q53GG8, Q6NSF4, Q8NHT8, Q9H3T4, Q9H4C6 | Gene names | GBA3, CBG, CBGL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic beta-glucosidase (EC 3.2.1.21) (Cytosolic beta-glucosidase- like protein 1). | |||||
|
GCP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.014392 (rank : 116) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
KV2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.002932 (rank : 137) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01627 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain V-II region VKappa167 precursor. | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.006483 (rank : 129) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
STX7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.007341 (rank : 128) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15400, Q5SZW2, Q96ES9 | Gene names | STX7 | |||
|
Domain Architecture |
|
|||||
| Description | Syntaxin-7. | |||||
|
STX7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.009177 (rank : 126) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70439 | Gene names | Stx7, Syn7 | |||
|
Domain Architecture |
|
|||||
| Description | Syntaxin-7. | |||||
|
TDRD7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.006217 (rank : 130) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | -0.005827 (rank : 156) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.001144 (rank : 144) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ZN583_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | -0.004419 (rank : 154) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V080, Q3V3L4 | Gene names | Znf583, Zfp583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583. | |||||
|
ZNF85_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | -0.004544 (rank : 155) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03923 | Gene names | ZNF85 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 85 (Zinc finger protein HPF4) (HTF1). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.063459 (rank : 77) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.072601 (rank : 76) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.392854 (rank : 73) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.405418 (rank : 71) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.051101 (rank : 83) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
UBP54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.055938 (rank : 81) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.055632 (rank : 82) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 149 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.995727 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.876781 (rank : 3) | θ value | 1.41672e-45 (rank : 3) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.861892 (rank : 4) | θ value | 4.41421e-39 (rank : 4) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.856857 (rank : 5) | θ value | 2.50655e-34 (rank : 6) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.839317 (rank : 6) | θ value | 5.96599e-36 (rank : 5) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.832109 (rank : 7) | θ value | 1.52067e-31 (rank : 8) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.828521 (rank : 8) | θ value | 3.50572e-28 (rank : 14) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.823503 (rank : 9) | θ value | 6.82597e-32 (rank : 7) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.790106 (rank : 10) | θ value | 2.69047e-20 (rank : 15) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.787479 (rank : 11) | θ value | 8.10077e-17 (rank : 17) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.787479 (rank : 12) | θ value | 8.10077e-17 (rank : 18) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.771259 (rank : 13) | θ value | 8.3442e-30 (rank : 11) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.771126 (rank : 14) | θ value | 1.2049e-28 (rank : 13) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.763255 (rank : 15) | θ value | 9.90251e-15 (rank : 24) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.763105 (rank : 16) | θ value | 3.3877e-31 (rank : 9) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.758303 (rank : 17) | θ value | 1.02238e-19 (rank : 16) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.754386 (rank : 18) | θ value | 5.40856e-29 (rank : 12) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.753572 (rank : 19) | θ value | 4.89182e-30 (rank : 10) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.741748 (rank : 20) | θ value | 4.44505e-15 (rank : 21) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.731624 (rank : 21) | θ value | 2.60593e-15 (rank : 20) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.730253 (rank : 22) | θ value | 4.44505e-15 (rank : 22) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.729509 (rank : 23) | θ value | 7.58209e-15 (rank : 23) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.718962 (rank : 24) | θ value | 1.80466e-16 (rank : 19) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.712767 (rank : 25) | θ value | 7.09661e-13 (rank : 27) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.711655 (rank : 26) | θ value | 1.47974e-10 (rank : 36) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.711444 (rank : 27) | θ value | 9.26847e-13 (rank : 28) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.711258 (rank : 28) | θ value | 7.84624e-12 (rank : 34) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.706532 (rank : 29) | θ value | 7.84624e-12 (rank : 33) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.704580 (rank : 30) | θ value | 1.58096e-12 (rank : 29) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.704145 (rank : 31) | θ value | 6.64225e-11 (rank : 35) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.704101 (rank : 32) | θ value | 6.41864e-14 (rank : 26) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.702761 (rank : 33) | θ value | 3.52202e-12 (rank : 31) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.685327 (rank : 34) | θ value | 1.17247e-07 (rank : 46) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.683049 (rank : 35) | θ value | 3.64472e-09 (rank : 39) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.681379 (rank : 36) | θ value | 1.9326e-10 (rank : 37) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.674140 (rank : 37) | θ value | 2.36244e-08 (rank : 41) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.669239 (rank : 38) | θ value | 1.29331e-14 (rank : 25) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.666946 (rank : 39) | θ value | 2.36244e-08 (rank : 43) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.666238 (rank : 40) | θ value | 2.36244e-08 (rank : 42) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.652918 (rank : 41) | θ value | 1.58096e-12 (rank : 30) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.646367 (rank : 42) | θ value | 3.52202e-12 (rank : 32) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.635695 (rank : 43) | θ value | 2.88788e-06 (rank : 56) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.627468 (rank : 44) | θ value | 0.00175202 (rank : 68) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.626355 (rank : 45) | θ value | 3.08544e-08 (rank : 44) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.599430 (rank : 46) | θ value | 6.87365e-08 (rank : 45) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.594966 (rank : 47) | θ value | 1.69304e-06 (rank : 55) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.593511 (rank : 48) | θ value | 1.29631e-06 (rank : 53) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.593485 (rank : 49) | θ value | 1.29631e-06 (rank : 52) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.590076 (rank : 50) | θ value | 6.43352e-06 (rank : 58) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.589904 (rank : 51) | θ value | 3.77169e-06 (rank : 57) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.581687 (rank : 52) | θ value | 2.44474e-05 (rank : 60) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.580835 (rank : 53) | θ value | 1.29631e-06 (rank : 54) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.576747 (rank : 54) | θ value | 0.00175202 (rank : 69) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.575642 (rank : 55) | θ value | 0.000602161 (rank : 67) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.565076 (rank : 56) | θ value | 1.53129e-07 (rank : 49) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.540227 (rank : 57) | θ value | 0.000461057 (rank : 64) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.538127 (rank : 58) | θ value | 0.00390308 (rank : 70) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.508630 (rank : 59) | θ value | 1.53129e-07 (rank : 48) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.505165 (rank : 60) | θ value | 3.41135e-07 (rank : 51) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.500159 (rank : 61) | θ value | 5.62301e-10 (rank : 38) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.497089 (rank : 62) | θ value | 3.64472e-09 (rank : 40) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.494888 (rank : 63) | θ value | 1.53129e-07 (rank : 47) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.491565 (rank : 64) | θ value | 1.99992e-07 (rank : 50) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.491486 (rank : 65) | θ value | 2.44474e-05 (rank : 59) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.484494 (rank : 66) | θ value | 0.000158464 (rank : 62) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.476370 (rank : 67) | θ value | 4.1701e-05 (rank : 61) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.469074 (rank : 68) | θ value | 0.000461057 (rank : 65) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.459603 (rank : 69) | θ value | 0.00035302 (rank : 63) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.436038 (rank : 70) | θ value | 0.0961366 (rank : 75) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.405418 (rank : 71) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.398724 (rank : 72) | θ value | 0.000602161 (rank : 66) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.392854 (rank : 73) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
ASXL1_MOUSE
|
||||||
| NC score | 0.081682 (rank : 74) | θ value | 0.0330416 (rank : 71) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.080225 (rank : 75) | θ value | 0.0431538 (rank : 72) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.072601 (rank : 76) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.063459 (rank : 77) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
SPT6H_HUMAN
|
||||||
| NC score | 0.063136 (rank : 78) | θ value | 0.163984 (rank : 77) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7KZ85, Q15737, Q6GMQ4, Q7KYW9, Q7LDK4, Q8N526, Q92775, Q96AH3, Q9BTH9, Q9BTI2 | Gene names | SUPT6H, KIAA0162, SPT6H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6 (hSPT6) (Tat-cotransactivator 2 protein) (Tat-CT2 protein). | |||||
|
SPT6H_MOUSE
|
||||||
| NC score | 0.062651 (rank : 79) | θ value | 0.163984 (rank : 78) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.062432 (rank : 80) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
UBP54_HUMAN
|
||||||
| NC score | 0.055938 (rank : 81) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q70EL1, Q5F2F4 | Gene names | USP54, C10orf29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP54_MOUSE
|
||||||
| NC score | 0.055632 (rank : 82) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BL06, Q8BZ28 | Gene names | Usp54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 54 (Inactive ubiquitin- specific peptidase 54). | |||||
|
UBP53_HUMAN
|
||||||
| NC score | 0.051101 (rank : 83) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70EK8, Q68DA5, Q8WVQ5, Q9P2J7 | Gene names | USP53, KIAA1350 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.049679 (rank : 84) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.048053 (rank : 85) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.043906 (rank : 86) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.043821 (rank : 87) | θ value | 0.21417 (rank : 79) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
TREX2_HUMAN
|
||||||
| NC score | 0.043498 (rank : 88) | θ value | 0.47712 (rank : 82) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQ50, Q45F08, Q9UN77 | Gene names | TREX2 | |||
|
Domain Architecture |
|
|||||
| Description | Three prime repair exonuclease 2 (EC 3.1.11.2) (3'-5' exonuclease TREX2). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.043231 (rank : 89) | θ value | 0.62314 (rank : 83) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
SAFB1_HUMAN
|
||||||
| NC score | 0.037302 (rank : 90) | θ value | 0.0961366 (rank : 74) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.036054 (rank : 91) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.035620 (rank : 92) | θ value | 0.0431538 (rank : 73) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.034728 (rank : 93) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.028810 (rank : 94) | θ value | 0.813845 (rank : 86) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
SQSTM_MOUSE
|
||||||
| NC score | 0.027977 (rank : 95) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
TREX2_MOUSE
|
||||||
| NC score | 0.027687 (rank : 96) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1A9 | Gene names | Trex2 | |||
|
Domain Architecture |
|
|||||
| Description | Three prime repair exonuclease 2 (EC 3.1.11.2) (3'-5' exonuclease TREX2). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.027082 (rank : 97) | θ value | 0.125558 (rank : 76) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CT172_MOUSE
|
||||||
| NC score | 0.024987 (rank : 98) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CYC5, Q9CYF2, Q9DA26 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf172 homolog. | |||||
|
FBX30_HUMAN
|
||||||
| NC score | 0.023488 (rank : 99) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TB52, Q9BXZ7 | Gene names | FBXO30, FBX30 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 30. | |||||
|
N4BP2_HUMAN
|
||||||
| NC score | 0.022451 (rank : 100) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.022298 (rank : 101) | θ value | 0.813845 (rank : 85) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
STON1_MOUSE
|
||||||
| NC score | 0.022233 (rank : 102) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
GAGC1_HUMAN
|
||||||
| NC score | 0.022004 (rank : 103) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60829, Q6IBI1 | Gene names | PAGE4, GAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen family C member 1 (Prostate-associated gene 4 protein) (PAGE-4) (PAGE-1) (GAGE-9). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.021950 (rank : 104) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.021464 (rank : 105) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
DBF4_MOUSE
|
||||||
| NC score | 0.020898 (rank : 106) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ41, Q3TQP4, Q3V1K2, Q6NSQ1, Q9CXF2 | Gene names | Dbf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (MuDBF4). | |||||
|
DLG4_MOUSE
|
||||||
| NC score | 0.018164 (rank : 107) | θ value | 0.47712 (rank : 81) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
DLG4_HUMAN
|
||||||
| NC score | 0.018125 (rank : 108) | θ value | 0.47712 (rank : 80) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.018104 (rank : 109) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.017329 (rank : 110) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
DLG1_MOUSE
|
||||||
| NC score | 0.017288 (rank : 111) | θ value | 1.06291 (rank : 87) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
DLG1_HUMAN
|
||||||
| NC score | 0.016874 (rank : 112) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
BNIP2_MOUSE
|
||||||
| NC score | 0.016699 (rank : 113) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54940 | Gene names | Bnip2, Nip2l | |||
|
Domain Architecture |

|
|||||
| Description | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2. | |||||
|
TOP3A_MOUSE
|
||||||
| NC score | 0.016057 (rank : 114) | θ value | 1.38821 (rank : 95) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70157 | Gene names | Top3a, Top3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 3-alpha (EC 5.99.1.2) (DNA topoisomerase III alpha). | |||||
|
GSTT2_MOUSE
|
||||||
| NC score | 0.015438 (rank : 115) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61133, Q61134, Q64472 | Gene names | Gstt2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
GCP5_HUMAN
|
||||||
| NC score | 0.014392 (rank : 116) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
SGCG_HUMAN
|
||||||
| NC score | 0.013537 (rank : 117) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13326 | Gene names | SGCG | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-sarcoglycan (Gamma-SG) (35 kDa dystrophin-associated glycoprotein) (35DAG). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.013392 (rank : 118) | θ value | 1.06291 (rank : 90) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.012828 (rank : 119) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
MGR4_HUMAN
|
||||||
| NC score | 0.011176 (rank : 120) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.010701 (rank : 121) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.010481 (rank : 122) | θ value | 0.813845 (rank : 84) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.010200 (rank : 123) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
MYEF2_HUMAN
|
||||||
| NC score | 0.010160 (rank : 124) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2K5, Q6NUM5, Q7L388, Q7Z4B7, Q9H922, Q9NUQ1 | Gene names | MYEF2, KIAA1341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin expression factor 2 (MyEF-2) (MST156). | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.010126 (rank : 125) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
STX7_MOUSE
|
||||||
| NC score | 0.009177 (rank : 126) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70439 | Gene names | Stx7, Syn7 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-7. | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.007521 (rank : 127) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
STX7_HUMAN
|
||||||
| NC score | 0.007341 (rank : 128) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15400, Q5SZW2, Q96ES9 | Gene names | STX7 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-7. | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.006483 (rank : 129) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
TDRD7_HUMAN
|
||||||
| NC score | 0.006217 (rank : 130) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TRAF5_HUMAN
|
||||||
| NC score | 0.005462 (rank : 131) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00463 | Gene names | TRAF5, RNF84 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5 (RING finger protein 84). | |||||
|
KV2E_MOUSE
|
||||||
| NC score | 0.005197 (rank : 132) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P03976 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region 17S29.1. | |||||
|
GBA3_HUMAN
|
||||||
| NC score | 0.004826 (rank : 133) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H227, Q3MIH4, Q53GG8, Q6NSF4, Q8NHT8, Q9H3T4, Q9H4C6 | Gene names | GBA3, CBG, CBGL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic beta-glucosidase (EC 3.2.1.21) (Cytosolic beta-glucosidase- like protein 1). | |||||
|
KV2A_MOUSE
|
||||||
| NC score | 0.003347 (rank : 134) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01626 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig kappa chain V-II region MOPC 167. | |||||
|
ATL2_MOUSE
|
||||||
| NC score | 0.003339 (rank : 135) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSK7, Q3U0C8 | Gene names | Adamtsl2, Kiaa0605, Tcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-like protein 2 precursor (ADAMTSL-2) (TSP1-repeats-containing protein 1) (TCP-1). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.003337 (rank : 136) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
KV2B_MOUSE
|
||||||
| NC score | 0.002932 (rank : 137) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01627 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain V-II region VKappa167 precursor. | |||||
|
ZPI_MOUSE
|
||||||
| NC score | 0.002866 (rank : 138) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R121, Q8VCV8 | Gene names | Serpina10, Zpi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Z-dependent protease inhibitor precursor (PZ-dependent protease inhibitor) (PZI) (Serpin A10). | |||||
|
K1H2_MOUSE
|
||||||
| NC score | 0.002851 (rank : 139) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
K1H1_MOUSE
|
||||||
| NC score | 0.002127 (rank : 140) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61765 | Gene names | Krtha1, Hka1, Krt1-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha1 (Hair keratin, type I Ha1) (HKA-1). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.002108 (rank : 141) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
K1H2_HUMAN
|
||||||
| NC score | 0.002010 (rank : 142) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.001745 (rank : 143) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.001144 (rank : 144) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.001089 (rank : 145) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.000081 (rank : 146) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
ZN518_HUMAN
|
||||||
| NC score | 0.000073 (rank : 147) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6AHZ1, O15044, Q32MP4 | Gene names | ZNF518, KIAA0335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 518. | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.000009 (rank : 148) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | -0.001472 (rank : 149) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
NEK8_MOUSE
|
||||||
| NC score | -0.003106 (rank : 150) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZR4, Q3U498, Q9D685 | Gene names | Nek8, Jck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8). | |||||
|
ZN583_HUMAN
|
||||||
| NC score | -0.003417 (rank : 151) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96ND8, O14850, Q2NKK3 | Gene names | ZNF583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583 (Zinc finger protein L3-5). | |||||
|
NEK8_HUMAN
|
||||||
| NC score | -0.003436 (rank : 152) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SG6, Q2M1S6, Q8NDH1 | Gene names | NEK8, JCK, NEK12A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek8 (EC 2.7.11.1) (Never in mitosis A-related kinase 8) (NimA-related protein kinase 8) (NIMA-related kinase 12a). | |||||
|
ZN431_HUMAN
|
||||||
| NC score | -0.004100 (rank : 153) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TF32, Q8IWC4 | Gene names | ZNF431, KIAA1969 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 431. | |||||
|
ZN583_MOUSE
|
||||||
| NC score | -0.004419 (rank : 154) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V080, Q3V3L4 | Gene names | Znf583, Zfp583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583. | |||||
|
ZNF85_HUMAN
|
||||||
| NC score | -0.004544 (rank : 155) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03923 | Gene names | ZNF85 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 85 (Zinc finger protein HPF4) (HTF1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.005827 (rank : 156) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 149 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||