Please be patient as the page loads
|
UBP26_MOUSE
|
||||||
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP26_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 5.07123e-136 (rank : 2) | NC score | 0.946074 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 7.87842e-113 (rank : 3) | NC score | 0.936398 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 7.42547e-87 (rank : 4) | NC score | 0.933426 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 2.31364e-80 (rank : 5) | NC score | 0.887991 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 6) | NC score | 0.507560 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 7) | NC score | 0.531119 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 8) | NC score | 0.552261 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 9) | NC score | 0.548036 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 10) | NC score | 0.548036 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 11) | NC score | 0.549722 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 12) | NC score | 0.516695 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 13) | NC score | 0.551084 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 14) | NC score | 0.501762 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 15) | NC score | 0.505566 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 16) | NC score | 0.343076 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 17) | NC score | 0.519456 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 18) | NC score | 0.505674 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 19) | NC score | 0.473679 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 20) | NC score | 0.515304 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 21) | NC score | 0.388244 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 22) | NC score | 0.398724 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 23) | NC score | 0.436234 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 24) | NC score | 0.446821 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 25) | NC score | 0.433026 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 26) | NC score | 0.437934 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 27) | NC score | 0.521437 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 28) | NC score | 0.428038 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 29) | NC score | 0.429308 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 30) | NC score | 0.431573 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 31) | NC score | 0.497742 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 32) | NC score | 0.462905 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 33) | NC score | 0.399041 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 34) | NC score | 0.445132 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 35) | NC score | 0.347276 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 36) | NC score | 0.371746 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP25_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 37) | NC score | 0.308053 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 38) | NC score | 0.326250 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 39) | NC score | 0.419364 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 40) | NC score | 0.308454 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 41) | NC score | 0.365985 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 42) | NC score | 0.420557 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 43) | NC score | 0.332182 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 44) | NC score | 0.425987 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 45) | NC score | 0.341169 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 46) | NC score | 0.417964 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
MAGD1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.023449 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.374880 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.400863 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.464885 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 0.163984 (rank : 51) | NC score | 0.465482 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 0.163984 (rank : 52) | NC score | 0.392587 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 0.21417 (rank : 53) | NC score | 0.421537 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.371481 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.435478 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 56) | NC score | 0.391506 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.379558 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
AKAP4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.030701 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60662, Q60630, Q8R2G7 | Gene names | Akap4, Akap82, Fsc1 | |||
|
Domain Architecture |
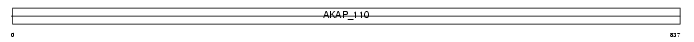
|
|||||
| Description | A-kinase anchor protein 4 precursor (Protein kinase A-anchoring protein 4) (PRKA4) (A-kinase anchor protein 82 kDa) (AKAP 82) (mAKAP82) (Major sperm fibrous sheath protein) (FSC1) (p82). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.010432 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
LIMA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.024865 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.013161 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.361225 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.363982 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.402813 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.010603 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
OSTP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.033466 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.360653 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
ZN454_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | -0.000821 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N9F8 | Gene names | ZNF454 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 454. | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.022686 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CF113_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.019686 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3T9Z9, Q5FWB1, Q6NXI1, Q8BJZ1, Q8BK40, Q8BKH3, Q8BL75, Q9CSP5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein C6orf113 homolog. | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.007307 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.359436 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.026146 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.010246 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.013369 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.012403 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.294008 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.306687 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
SYCP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.027505 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.011587 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.397365 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.399886 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
ZN208_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | -0.001076 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43345 | Gene names | ZNF208, ZNF91L | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 208. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.007907 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.350187 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.376533 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
ZNF96_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | -0.001262 (rank : 109) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 869 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43309, O43724 | Gene names | ZNF96, KIAA0426, ZNF305, ZSCAN12 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 96 (Zinc finger protein 305) (Zinc finger and SCAN domain-containing protein 12). | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.017418 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.004024 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
RPGF4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.017570 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.375077 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.373467 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.011062 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
ORC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.014027 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88708, Q9QYX1 | Gene names | Orc4l, Orc4 | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 4. | |||||
|
RBGPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.012379 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2M9, O75872, Q9HAB0, Q9UFJ7, Q9UQ15 | Gene names | RAB3GAP2, KIAA0839 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein non-catalytic subunit (Rab3 GTPase- activating protein 150 kDa subunit) (Rab3-GAP p150) (Rab3-GAP regulatory subunit) (RAB3-GAP150) (RGAP-iso). | |||||
|
SETB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.012328 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | -0.000353 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.413937 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
CC062_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.013355 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZUJ4, Q6P7E9, Q7Z3X6 | Gene names | C3orf62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf62. | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.005664 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.006103 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
PDIA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.008127 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13667 | Gene names | PDIA4, ERP70, ERP72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
PDIA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.008154 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08003, P15841 | Gene names | Pdia4, Cai, Erp72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
RPGF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.010077 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
SENP7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.011577 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQF6, Q96PS5, Q9C0F6, Q9HBT5 | Gene names | SENP7, KIAA1707, SSP2, SUSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 7 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP7) (SUMO-1-specific protease 2). | |||||
|
SPAT7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.011792 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0W8, Q8WX30, Q96HF3, Q9H0X0, Q9P0W7 | Gene names | SPATA7, HSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 (Spermatogenesis-associated protein HSD3) (HSD-3.1). | |||||
|
ZN274_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | -0.001817 (rank : 110) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GC6, Q8WY37, Q8WY38, Q92969, Q9UII0, Q9UII1 | Gene names | ZNF274 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 274 (Zinc finger protein HFB101) (Zinc finger protein zfp2) (Zf2). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.296546 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.294343 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.327145 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.946074 (rank : 2) | θ value | 5.07123e-136 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.936398 (rank : 3) | θ value | 7.87842e-113 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.933426 (rank : 4) | θ value | 7.42547e-87 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.887991 (rank : 5) | θ value | 2.31364e-80 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.552261 (rank : 6) | θ value | 3.29651e-10 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.551084 (rank : 7) | θ value | 1.99992e-07 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.549722 (rank : 8) | θ value | 6.87365e-08 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.548036 (rank : 9) | θ value | 6.21693e-09 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.548036 (rank : 10) | θ value | 6.21693e-09 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.531119 (rank : 11) | θ value | 1.47974e-10 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.521437 (rank : 12) | θ value | 0.00102713 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.519456 (rank : 13) | θ value | 8.40245e-06 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.516695 (rank : 14) | θ value | 1.17247e-07 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.515304 (rank : 15) | θ value | 7.1131e-05 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.507560 (rank : 16) | θ value | 5.08577e-11 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.505674 (rank : 17) | θ value | 2.44474e-05 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.505566 (rank : 18) | θ value | 1.29631e-06 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.501762 (rank : 19) | θ value | 1.99992e-07 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.497742 (rank : 20) | θ value | 0.00175202 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.473679 (rank : 21) | θ value | 2.44474e-05 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.465482 (rank : 22) | θ value | 0.163984 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.464885 (rank : 23) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.462905 (rank : 24) | θ value | 0.00298849 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.446821 (rank : 25) | θ value | 0.000786445 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.445132 (rank : 26) | θ value | 0.00509761 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.437934 (rank : 27) | θ value | 0.00102713 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.436234 (rank : 28) | θ value | 0.000786445 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.435478 (rank : 29) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.433026 (rank : 30) | θ value | 0.00102713 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.431573 (rank : 31) | θ value | 0.00134147 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.429308 (rank : 32) | θ value | 0.00134147 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.428038 (rank : 33) | θ value | 0.00134147 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.425987 (rank : 34) | θ value | 0.0252991 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.421537 (rank : 35) | θ value | 0.21417 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.420557 (rank : 36) | θ value | 0.0193708 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.419364 (rank : 37) | θ value | 0.0113563 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.417964 (rank : 38) | θ value | 0.0330416 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.413937 (rank : 39) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.402813 (rank : 40) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.400863 (rank : 41) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.399886 (rank : 42) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.399041 (rank : 43) | θ value | 0.00298849 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.398724 (rank : 44) | θ value | 0.000602161 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.397365 (rank : 45) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.392587 (rank : 46) | θ value | 0.163984 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.391506 (rank : 47) | θ value | 0.279714 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.388244 (rank : 48) | θ value | 0.000461057 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.379558 (rank : 49) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.376533 (rank : 50) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.375077 (rank : 51) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.374880 (rank : 52) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.373467 (rank : 53) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.371746 (rank : 54) | θ value | 0.00665767 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.371481 (rank : 55) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.365985 (rank : 56) | θ value | 0.0148317 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.363982 (rank : 57) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.361225 (rank : 58) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.360653 (rank : 59) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.359436 (rank : 60) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.350187 (rank : 61) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.347276 (rank : 62) | θ value | 0.00509761 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.343076 (rank : 63) | θ value | 6.43352e-06 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.341169 (rank : 64) | θ value | 0.0252991 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.332182 (rank : 65) | θ value | 0.0193708 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.327145 (rank : 66) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.326250 (rank : 67) | θ value | 0.0113563 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.308454 (rank : 68) | θ value | 0.0148317 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.308053 (rank : 69) | θ value | 0.00869519 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.306687 (rank : 70) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.296546 (rank : 71) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.294343 (rank : 72) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.294008 (rank : 73) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
OSTP_MOUSE
|
||||||
| NC score | 0.033466 (rank : 74) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
AKAP4_MOUSE
|
||||||
| NC score | 0.030701 (rank : 75) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60662, Q60630, Q8R2G7 | Gene names | Akap4, Akap82, Fsc1 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 4 precursor (Protein kinase A-anchoring protein 4) (PRKA4) (A-kinase anchor protein 82 kDa) (AKAP 82) (mAKAP82) (Major sperm fibrous sheath protein) (FSC1) (p82). | |||||
|
SYCP2_HUMAN
|
||||||
| NC score | 0.027505 (rank : 76) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BX26, O75763, Q5JX11, Q9NTX8, Q9UG27 | Gene names | SYCP2, SCP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptonemal complex protein 2 (SCP-2) (Synaptonemal complex lateral element protein) (hsSCP2). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.026146 (rank : 77) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
LIMA1_HUMAN
|
||||||
| NC score | 0.024865 (rank : 78) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
MAGD1_MOUSE
|
||||||
| NC score | 0.023449 (rank : 79) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QYH6, Q543L6, Q99PB5, Q9CYX1 | Gene names | Maged1, Nrage | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (Dlxin-1). | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.022686 (rank : 80) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CF113_MOUSE
|
||||||
| NC score | 0.019686 (rank : 81) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3T9Z9, Q5FWB1, Q6NXI1, Q8BJZ1, Q8BK40, Q8BKH3, Q8BL75, Q9CSP5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein C6orf113 homolog. | |||||
|
RPGF4_MOUSE
|
||||||
| NC score | 0.017570 (rank : 82) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQZ6, Q8VIP9, Q9CW52, Q9Z1P0 | Gene names | Rapgef4, Cgef2, Epac2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2) (cAMP-dependent Rap1 guanine-nucleotide exchange factor). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.017418 (rank : 83) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
ORC4_MOUSE
|
||||||
| NC score | 0.014027 (rank : 84) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88708, Q9QYX1 | Gene names | Orc4l, Orc4 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 4. | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.013369 (rank : 85) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
CC062_HUMAN
|
||||||
| NC score | 0.013355 (rank : 86) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZUJ4, Q6P7E9, Q7Z3X6 | Gene names | C3orf62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf62. | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.013161 (rank : 87) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.012403 (rank : 88) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
RBGPR_HUMAN
|
||||||
| NC score | 0.012379 (rank : 89) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2M9, O75872, Q9HAB0, Q9UFJ7, Q9UQ15 | Gene names | RAB3GAP2, KIAA0839 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein non-catalytic subunit (Rab3 GTPase- activating protein 150 kDa subunit) (Rab3-GAP p150) (Rab3-GAP regulatory subunit) (RAB3-GAP150) (RGAP-iso). | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.012328 (rank : 90) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SPAT7_HUMAN
|
||||||
| NC score | 0.011792 (rank : 91) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0W8, Q8WX30, Q96HF3, Q9H0X0, Q9P0W7 | Gene names | SPATA7, HSD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 (Spermatogenesis-associated protein HSD3) (HSD-3.1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.011587 (rank : 92) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
SENP7_HUMAN
|
||||||
| NC score | 0.011577 (rank : 93) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQF6, Q96PS5, Q9C0F6, Q9HBT5 | Gene names | SENP7, KIAA1707, SSP2, SUSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 7 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP7) (SUMO-1-specific protease 2). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.011062 (rank : 94) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.010603 (rank : 95) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.010432 (rank : 96) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.010246 (rank : 97) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
RPGF4_HUMAN
|
||||||
| NC score | 0.010077 (rank : 98) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WZA2, O95636, Q8IXK6, Q8TAA4 | Gene names | RAPGEF4, CGEF2, EPAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 4 (cAMP-regulated guanine nucleotide exchange factor II) (cAMP-GEFII) (Exchange factor directly activated by cAMP 2) (Epac 2). | |||||
|
PDIA4_MOUSE
|
||||||
| NC score | 0.008154 (rank : 99) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08003, P15841 | Gene names | Pdia4, Cai, Erp72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
PDIA4_HUMAN
|
||||||
| NC score | 0.008127 (rank : 100) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13667 | Gene names | PDIA4, ERP70, ERP72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.007907 (rank : 101) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.007307 (rank : 102) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.006103 (rank : 103) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.005664 (rank : 104) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.004024 (rank : 105) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | -0.000353 (rank : 106) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
ZN454_HUMAN
|
||||||
| NC score | -0.000821 (rank : 107) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N9F8 | Gene names | ZNF454 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 454. | |||||
|
ZN208_HUMAN
|
||||||
| NC score | -0.001076 (rank : 108) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43345 | Gene names | ZNF208, ZNF91L | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 208. | |||||
|
ZNF96_HUMAN
|
||||||
| NC score | -0.001262 (rank : 109) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 869 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43309, O43724 | Gene names | ZNF96, KIAA0426, ZNF305, ZSCAN12 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 96 (Zinc finger protein 305) (Zinc finger and SCAN domain-containing protein 12). | |||||
|
ZN274_HUMAN
|
||||||
| NC score | -0.001817 (rank : 110) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96GC6, Q8WY37, Q8WY38, Q92969, Q9UII0, Q9UII1 | Gene names | ZNF274 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 274 (Zinc finger protein HFB101) (Zinc finger protein zfp2) (Zf2). | |||||