Please be patient as the page loads
|
UBP43_HUMAN
|
||||||
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBP31_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.942843 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.988926 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 6.81017e-40 (rank : 4) | NC score | 0.857166 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP4_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 5) | NC score | 0.856078 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP15_HUMAN
|
||||||
| θ value | 6.37413e-38 (rank : 6) | NC score | 0.854088 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP15_MOUSE
|
||||||
| θ value | 6.37413e-38 (rank : 7) | NC score | 0.854511 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 4.27553e-34 (rank : 8) | NC score | 0.855788 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 9) | NC score | 0.832944 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 1.6813e-30 (rank : 10) | NC score | 0.793422 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP2_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 11) | NC score | 0.847069 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP2_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 12) | NC score | 0.844702 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP50_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 13) | NC score | 0.842159 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 14) | NC score | 0.811551 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP33_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 15) | NC score | 0.822965 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 16) | NC score | 0.706191 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 17) | NC score | 0.748745 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP20_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 18) | NC score | 0.824524 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP21_HUMAN
|
||||||
| θ value | 4.9032e-22 (rank : 19) | NC score | 0.828309 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP21_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 20) | NC score | 0.829459 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP3_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 21) | NC score | 0.823917 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 22) | NC score | 0.823572 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP22_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 23) | NC score | 0.805200 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 24) | NC score | 0.806421 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP50_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 25) | NC score | 0.826640 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP44_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 26) | NC score | 0.761602 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP49_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 27) | NC score | 0.749594 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP49_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 28) | NC score | 0.747398 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 29) | NC score | 0.685486 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 30) | NC score | 0.686581 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 31) | NC score | 0.773659 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UB17L_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 32) | NC score | 0.729411 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP18_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 33) | NC score | 0.722212 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 34) | NC score | 0.700149 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP18_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 35) | NC score | 0.735157 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 36) | NC score | 0.712672 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP46_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 37) | NC score | 0.724816 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 38) | NC score | 0.724816 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBPW_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 39) | NC score | 0.746092 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP12_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 40) | NC score | 0.728510 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP38_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 41) | NC score | 0.603811 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
UBP38_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 42) | NC score | 0.605754 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 43) | NC score | 0.592203 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 44) | NC score | 0.537326 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 45) | NC score | 0.537470 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 46) | NC score | 0.609762 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 47) | NC score | 0.619385 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP30_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 48) | NC score | 0.680168 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP26_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 49) | NC score | 0.518552 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 50) | NC score | 0.493576 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 51) | NC score | 0.533682 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 52) | NC score | 0.610844 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 53) | NC score | 0.615127 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 54) | NC score | 0.541928 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 55) | NC score | 0.541006 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 56) | NC score | 0.581687 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP29_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 57) | NC score | 0.526217 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP14_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 58) | NC score | 0.555202 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP40_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 59) | NC score | 0.598792 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
UBP14_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 60) | NC score | 0.566684 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 61) | NC score | 0.584702 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
SNUT2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 62) | NC score | 0.595447 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
SNUT2_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 63) | NC score | 0.601635 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP7_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 64) | NC score | 0.589426 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 65) | NC score | 0.477255 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 66) | NC score | 0.472629 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 67) | NC score | 0.354022 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP29_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 68) | NC score | 0.488545 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
CI079_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 69) | NC score | 0.053086 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 70) | NC score | 0.608938 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 71) | NC score | 0.035246 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 72) | NC score | 0.038724 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 73) | NC score | 0.002597 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 74) | NC score | 0.037967 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
UBP28_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 75) | NC score | 0.349971 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP25_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 76) | NC score | 0.324816 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
UBP25_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 77) | NC score | 0.326257 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 78) | NC score | 0.039542 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
TM108_HUMAN
|
||||||
| θ value | 0.163984 (rank : 79) | NC score | 0.033881 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 0.21417 (rank : 80) | NC score | 0.012984 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 81) | NC score | 0.014503 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 82) | NC score | 0.022396 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 0.47712 (rank : 83) | NC score | 0.018861 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 84) | NC score | 0.031798 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 85) | NC score | 0.018948 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 86) | NC score | 0.024017 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 87) | NC score | 0.032870 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.038178 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.038314 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 90) | NC score | 0.033043 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 1.06291 (rank : 91) | NC score | 0.402813 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.004950 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CO2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.004937 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.024895 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 95) | NC score | 0.021255 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.020530 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.022771 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 1.38821 (rank : 98) | NC score | 0.009479 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.007641 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
MO4L2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.020092 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
MO4L2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.020213 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.026116 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | 0.014184 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.031812 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.016653 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TYDP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.016885 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ37, Q8VE39 | Gene names | Tdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosyl-DNA phosphodiesterase 1 (EC 3.1.4.-) (Tyr-DNA phosphodiesterase 1). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.038465 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
EPHB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | -0.002457 (rank : 150) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54753, Q7Z740 | Gene names | EPHB3, ETK2, HEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HEK-2). | |||||
|
EPHB3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | -0.002445 (rank : 149) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54754, Q62214 | Gene names | Ephb3, Etk2, Mdk5, Sek4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-5) (Developmental kinase 5) (SEK-4). | |||||
|
LAEVR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.006953 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
|
PACE1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.012323 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBQ7, Q6NY01, Q8BQC9, Q8BRJ1 | Gene names | Scyl3, Pace1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-associating with the carboxyl-terminal domain of ezrin (Ezrin- binding protein PACE-1) (SCY1-like protein 3). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.019426 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.002472 (rank : 146) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
DGKG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.007712 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.033154 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.007928 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.002815 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.018364 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.006229 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FOXK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.003545 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
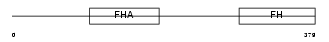
Domain Architecture |
|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
GLDN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.027853 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZMI3, Q6UXZ7, Q7Z359 | Gene names | GLDN, COLM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gliomedin. | |||||
|
K0310_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.015283 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.022392 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PMS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.009388 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
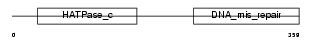
Domain Architecture |
|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | -0.001288 (rank : 148) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
CO4A6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.030322 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14031, Q12823, Q14053, Q5JYH6, Q9NQM5, Q9NTX3, Q9UJ76, Q9UMG6, Q9Y4L4 | Gene names | COL4A6 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-6(IV) chain precursor. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.014078 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.005984 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
KPBB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.006156 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93100, Q8N4T5 | Gene names | PHKB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit beta (Phosphorylase kinase subunit beta). | |||||
|
LAMP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.009884 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQV4, O94781, Q8NEC8 | Gene names | LAMP3, DCLAMP, TSC403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosome-associated membrane glycoprotein 3 precursor (LAMP-3) (Lysosomal-associated membrane protein 3) (DC-lysosome-associated membrane glycoprotein) (DC LAMP) (Protein TSC403) (CD208 antigen). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.006987 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
SGIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.021379 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.017236 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.030833 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | -0.002853 (rank : 151) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.001143 (rank : 147) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CT174_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.012236 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.004169 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.002491 (rank : 145) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.006524 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
PDIA5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.004428 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921X9 | Gene names | Pdia5, Pdir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase A5 precursor (EC 5.3.4.1) (Protein disulfide isomerase-related protein). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.006345 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
TICN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.003597 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
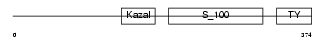
Domain Architecture |
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.006561 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.086556 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
BRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.098251 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
GGNB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.096275 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5YKI7, Q5YKI8 | Gene names | GGNBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
GGNB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.126792 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6K1E7, Q8C5T9 | Gene names | Ggnbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
HDAC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.062979 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
HDAC6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.067831 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
TBCD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.063071 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZP1, Q9H0B9, Q9UDD4 | Gene names | TBC1D3, PRC17 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 3 (Rab GTPase-activating protein PRC17) (Prostate cancer gene 17 protein) (TRE17 alpha protein). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.988926 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.942843 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.857166 (rank : 4) | θ value | 6.81017e-40 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
UBP4_MOUSE
|
||||||
| NC score | 0.856078 (rank : 5) | θ value | 1.16164e-39 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P35123, O54704 | Gene names | Usp4, Unp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.855788 (rank : 6) | θ value | 4.27553e-34 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
UBP15_MOUSE
|
||||||
| NC score | 0.854511 (rank : 7) | θ value | 6.37413e-38 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R5H1, Q3UL25 | Gene names | Usp15 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15). | |||||
|
UBP15_HUMAN
|
||||||
| NC score | 0.854088 (rank : 8) | θ value | 6.37413e-38 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y4E8, Q9HCA6, Q9UNP0, Q9Y5B5 | Gene names | USP15, KIAA0529 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 15 (EC 3.1.2.15) (Ubiquitin thioesterase 15) (Ubiquitin-specific-processing protease 15) (Deubiquitinating enzyme 15) (Unph-2) (Unph4). | |||||
|
UBP2_MOUSE
|
||||||
| NC score | 0.847069 (rank : 9) | θ value | 8.63488e-27 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88623, Q6X4T9, Q6X4U1, Q8VD74, Q9JJ87 | Gene names | Usp2, Ubp41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP2_HUMAN
|
||||||
| NC score | 0.844702 (rank : 10) | θ value | 4.28545e-26 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75604, Q96MB9, Q9BQ21 | Gene names | USP2, UBP41 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 2 (EC 3.1.2.15) (Ubiquitin thioesterase 2) (Ubiquitin-specific-processing protease 2) (Deubiquitinating enzyme 2) (41 kDa ubiquitin-specific protease). | |||||
|
UBP50_MOUSE
|
||||||
| NC score | 0.842159 (rank : 11) | θ value | 7.30988e-26 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P8X6, Q9D558, Q9D9E7 | Gene names | Usp50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ubiquitin carboxyl-terminal hydrolase 50 (EC 3.1.2.15) (Ubiquitin thioesterase 50) (Ubiquitin-specific-processing protease 50) (Deubiquitinating enzyme 50). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.832944 (rank : 12) | θ value | 7.54701e-31 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
UBP21_MOUSE
|
||||||
| NC score | 0.829459 (rank : 13) | θ value | 6.40375e-22 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QZL6, Q9D0R1 | Gene names | Usp21, Usp23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21). | |||||
|
UBP21_HUMAN
|
||||||
| NC score | 0.828309 (rank : 14) | θ value | 4.9032e-22 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UK80, Q9BTV1, Q9NYN4 | Gene names | USP21, USP23 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 21 (EC 3.1.2.15) (Ubiquitin thioesterase 21) (Ubiquitin-specific-processing protease 21) (Deubiquitinating enzyme 21) (NEDD8-specific protease). | |||||
|
UBP50_HUMAN
|
||||||
| NC score | 0.826640 (rank : 15) | θ value | 8.10077e-17 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q70EL3 | Gene names | USP50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 50 (Inactive ubiquitin- specific peptidase 50). | |||||
|
UBP20_HUMAN
|
||||||
| NC score | 0.824524 (rank : 16) | θ value | 5.23862e-24 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9Y2K6, Q96LG5, Q9UQN8, Q9UQP0 | Gene names | USP20, KIAA1003, LSFR3A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 20 (EC 3.1.2.15) (Ubiquitin thioesterase 20) (Ubiquitin-specific-processing protease 20) (Deubiquitinating enzyme 20). | |||||
|
UBP3_MOUSE
|
||||||
| NC score | 0.823917 (rank : 17) | θ value | 8.36355e-22 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91W36 | Gene names | Usp3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP3_HUMAN
|
||||||
| NC score | 0.823572 (rank : 18) | θ value | 1.86321e-21 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y6I4 | Gene names | USP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 3 (EC 3.1.2.15) (Ubiquitin thioesterase 3) (Ubiquitin-specific-processing protease 3) (Deubiquitinating enzyme 3). | |||||
|
UBP33_HUMAN
|
||||||
| NC score | 0.822965 (rank : 19) | θ value | 2.35151e-24 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8TEY7, Q8TEY6, Q96AV6, Q9H9F0, Q9UPQ5 | Gene names | USP33, KIAA1097, VDU1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 33 (EC 3.1.2.15) (Ubiquitin thioesterase 33) (Ubiquitin-specific-processing protease 33) (Deubiquitinating enzyme 33) (VHL-interacting deubiquitinating enzyme 1). | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.811551 (rank : 20) | θ value | 1.05554e-24 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
UBP22_MOUSE
|
||||||
| NC score | 0.806421 (rank : 21) | θ value | 5.42112e-21 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5DU02, Q5SU81, Q66JV8 | Gene names | Usp22, Kiaa1063 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP22_HUMAN
|
||||||
| NC score | 0.805200 (rank : 22) | θ value | 4.15078e-21 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPT9 | Gene names | USP22, KIAA1063, USP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 22 (EC 3.1.2.15) (Ubiquitin thioesterase 22) (Ubiquitin-specific-processing protease 22) (Deubiquitinating enzyme 22). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.793422 (rank : 23) | θ value | 1.6813e-30 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.773659 (rank : 24) | θ value | 1.9326e-10 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
UBP44_HUMAN
|
||||||
| NC score | 0.761602 (rank : 25) | θ value | 1.29331e-14 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9H0E7 | Gene names | USP44 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 44 (EC 3.1.2.15) (Ubiquitin thioesterase 44) (Ubiquitin-specific-processing protease 44) (Deubiquitinating enzyme 44). | |||||
|
UBP49_MOUSE
|
||||||
| NC score | 0.749594 (rank : 26) | θ value | 1.09485e-13 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q6P9L4 | Gene names | Usp49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.748745 (rank : 27) | θ value | 4.01107e-24 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
UBP49_HUMAN
|
||||||
| NC score | 0.747398 (rank : 28) | θ value | 1.86753e-13 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q70CQ1, Q96CK4 | Gene names | USP49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 49 (EC 3.1.2.15) (Ubiquitin thioesterase 49) (Ubiquitin-specific-processing protease 49) (Deubiquitinating enzyme 49). | |||||
|
UBPW_MOUSE
|
||||||
| NC score | 0.746092 (rank : 29) | θ value | 1.06045e-08 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q61068 | Gene names | Dub1, Dub-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase DUB-1 (EC 3.1.2.15) (Ubiquitin thioesterase DUB-1) (Ubiquitin-specific-processing protease DUB-1) (Deubiquitinating enzyme 1). | |||||
|
UBP18_HUMAN
|
||||||
| NC score | 0.735157 (rank : 30) | θ value | 1.25267e-09 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UMW8, Q9NY71 | Gene names | USP18, ISG43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease) (hUBP43). | |||||
|
UB17L_HUMAN
|
||||||
| NC score | 0.729411 (rank : 31) | θ value | 2.52405e-10 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q7RTZ2 | Gene names | USP17L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 17-like protein (EC 3.1.2.15) (Ubiquitin thioesterase 17-like) (Ubiquitin-specific-processing protease 17-like) (Deubiquitinating enzyme 17-like). | |||||
|
UBP12_HUMAN
|
||||||
| NC score | 0.728510 (rank : 32) | θ value | 2.36244e-08 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O75317 | Gene names | USP12, UBH1, USP12L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 12 (EC 3.1.2.15) (Ubiquitin thioesterase 12) (Ubiquitin-specific-processing protease 12) (Deubiquitinating enzyme 12) (Ubiquitin-hydrolyzing enzyme 1). | |||||
|
UBP46_HUMAN
|
||||||
| NC score | 0.724816 (rank : 33) | θ value | 6.21693e-09 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62068, Q80V95, Q9H7U4, Q9H9T8 | Gene names | USP46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP46_MOUSE
|
||||||
| NC score | 0.724816 (rank : 34) | θ value | 6.21693e-09 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P62069, Q80V95, Q9H7U4, Q9H9T8 | Gene names | Usp46 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 46 (EC 3.1.2.15) (Ubiquitin thioesterase 46) (Ubiquitin-specific-processing protease 46) (Deubiquitinating enzyme 46). | |||||
|
UBP18_MOUSE
|
||||||
| NC score | 0.722212 (rank : 35) | θ value | 5.62301e-10 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WTV6 | Gene names | Usp18, Ubp43 | |||
|
Domain Architecture |

|
|||||
| Description | Ubl carboxyl-terminal hydrolase 18 (EC 3.1.2.-) (Ubl thioesterase 18) (ISG15-specific-processing protease) (43 kDa ISG15-specific protease). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.712672 (rank : 36) | θ value | 6.21693e-09 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.706191 (rank : 37) | θ value | 3.07116e-24 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.700149 (rank : 38) | θ value | 7.34386e-10 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBP48_HUMAN
|
||||||
| NC score | 0.686581 (rank : 39) | θ value | 3.89403e-11 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q86UV5, Q2M3I4, Q5SZI4, Q5T3T5, Q6NX53, Q8N3F6, Q96F64, Q96IQ3, Q9H5N3, Q9H5T7, Q9NUJ6, Q9NXR0 | Gene names | USP48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.685486 (rank : 40) | θ value | 1.74796e-11 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
UBP30_HUMAN
|
||||||
| NC score | 0.680168 (rank : 41) | θ value | 3.77169e-06 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ3, Q8WTU7, Q96JX4, Q9BSS3 | Gene names | USP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 30 (EC 3.1.2.15) (Ubiquitin thioesterase 30) (Ubiquitin-specific-processing protease 30) (Deubiquitinating enzyme 30). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.619385 (rank : 42) | θ value | 2.88788e-06 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.615127 (rank : 43) | θ value | 1.43324e-05 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.610844 (rank : 44) | θ value | 1.43324e-05 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.609762 (rank : 45) | θ value | 1.29631e-06 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.608938 (rank : 46) | θ value | 0.0330416 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
UBP38_HUMAN
|
||||||
| NC score | 0.605754 (rank : 47) | θ value | 1.53129e-07 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8NB14, Q8NDF5, Q96DK6, Q96PZ6, Q9BY55 | Gene names | USP38, KIAA1891 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38) (HP43.8KD). | |||||
|
UBP38_MOUSE
|
||||||
| NC score | 0.603811 (rank : 48) | θ value | 5.26297e-08 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BW70, Q8BWL1, Q8BX03 | Gene names | Usp38 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 38 (EC 3.1.2.15) (Ubiquitin thioesterase 38) (Ubiquitin-specific-processing protease 38) (Deubiquitinating enzyme 38). | |||||
|
SNUT2_MOUSE
|
||||||
| NC score | 0.601635 (rank : 49) | θ value | 0.000121331 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q3TIX9, Q3TI55, Q8BLI5, Q8BZZ5, Q9CQR1 | Gene names | Usp39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (Inactive ubiquitin-specific peptidase 39). | |||||
|
UBP40_HUMAN
|
||||||
| NC score | 0.598792 (rank : 50) | θ value | 7.1131e-05 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NVE5, Q6NX38, Q70EL0 | Gene names | USP40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 40 (EC 3.1.2.15) (Ubiquitin thioesterase 40) (Ubiquitin-specific-processing protease 40) (Deubiquitinating enzyme 40). | |||||
|
SNUT2_HUMAN
|
||||||
| NC score | 0.595447 (rank : 51) | θ value | 0.000121331 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q53GS9, Q6NX47, Q96RK9, Q9BV89, Q9H381, Q9P050, Q9Y310 | Gene names | USP39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 2 (U4/U6.U5 tri-snRNP-associated 65 kDa protein) (65K) (Inactive ubiquitin-specific peptidase 39) (SAD1 homolog). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.592203 (rank : 52) | θ value | 3.41135e-07 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
UBP7_HUMAN
|
||||||
| NC score | 0.589426 (rank : 53) | θ value | 0.00134147 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q93009 | Gene names | USP7, HAUSP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 7 (EC 3.1.2.15) (Ubiquitin thioesterase 7) (Ubiquitin-specific-processing protease 7) (Deubiquitinating enzyme 7) (Herpesvirus-associated ubiquitin-specific protease). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.584702 (rank : 54) | θ value | 9.29e-05 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.581687 (rank : 55) | θ value | 2.44474e-05 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP14_HUMAN
|
||||||
| NC score | 0.566684 (rank : 56) | θ value | 9.29e-05 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P54578 | Gene names | USP14, TGT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP14_MOUSE
|
||||||
| NC score | 0.555202 (rank : 57) | θ value | 7.1131e-05 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JMA1, Q543U5, Q923F2, Q9D0L0 | Gene names | Usp14 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 14 (EC 3.1.2.15) (Ubiquitin thioesterase 14) (Ubiquitin-specific-processing protease 14) (Deubiquitinating enzyme 14). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.541928 (rank : 58) | θ value | 1.87187e-05 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.541006 (rank : 59) | θ value | 1.87187e-05 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.537470 (rank : 60) | θ value | 7.59969e-07 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.537326 (rank : 61) | θ value | 4.45536e-07 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.533682 (rank : 62) | θ value | 1.09739e-05 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
UBP29_HUMAN
|
||||||
| NC score | 0.526217 (rank : 63) | θ value | 3.19293e-05 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9HBJ7 | Gene names | USP29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP26_HUMAN
|
||||||
| NC score | 0.518552 (rank : 64) | θ value | 6.43352e-06 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BXU7 | Gene names | USP26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.493576 (rank : 65) | θ value | 8.40245e-06 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
UBP29_MOUSE
|
||||||
| NC score | 0.488545 (rank : 66) | θ value | 0.00869519 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9ES63 | Gene names | Usp29 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 29 (EC 3.1.2.15) (Ubiquitin thioesterase 29) (Ubiquitin-specific-processing protease 29) (Deubiquitinating enzyme 29). | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.477255 (rank : 67) | θ value | 0.00228821 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP5_MOUSE
|
||||||
| NC score | 0.472629 (rank : 68) | θ value | 0.00228821 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P56399 | Gene names | Usp5, Isot | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.402813 (rank : 69) | θ value | 1.06291 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.354022 (rank : 70) | θ value | 0.00298849 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP28_HUMAN
|
||||||
| NC score | 0.349971 (rank : 71) | θ value | 0.0563607 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96RU2, Q9P213 | Gene names | USP28, KIAA1515 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
UBP25_MOUSE
|
||||||
| NC score | 0.326257 (rank : 72) | θ value | 0.0736092 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P57080, Q80ZT9 | Gene names | Usp25 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (mUSP25). | |||||
|
UBP25_HUMAN
|
||||||
| NC score | 0.324816 (rank : 73) | θ value | 0.0736092 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHP3, Q9H9W1 | Gene names | USP25, USP21 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 25 (EC 3.1.2.15) (Ubiquitin thioesterase 25) (Ubiquitin-specific-processing protease 25) (Deubiquitinating enzyme 25) (USP on chromosome 21). | |||||
|
GGNB1_MOUSE
|
||||||
| NC score | 0.126792 (rank : 74) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6K1E7, Q8C5T9 | Gene names | Ggnbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
BRAP_MOUSE
|
||||||
| NC score | 0.098251 (rank : 75) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MP8, Q8CC00, Q8CHX1, Q99MP7, Q9CXX8 | Gene names | Brap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP). | |||||
|
GGNB1_HUMAN
|
||||||
| NC score | 0.096275 (rank : 76) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5YKI7, Q5YKI8 | Gene names | GGNBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin-binding protein 1. | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.086556 (rank : 77) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
HDAC6_MOUSE
|
||||||
| NC score | 0.067831 (rank : 78) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z2V5 | Gene names | Hdac6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6) (Histone deacetylase mHDA2). | |||||
|
TBCD3_HUMAN
|
||||||
| NC score | 0.063071 (rank : 79) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZP1, Q9H0B9, Q9UDD4 | Gene names | TBC1D3, PRC17 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 3 (Rab GTPase-activating protein PRC17) (Prostate cancer gene 17 protein) (TRE17 alpha protein). | |||||
|
HDAC6_HUMAN
|
||||||
| NC score | 0.062979 (rank : 80) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBN7 | Gene names | HDAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 6 (HD6). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.053086 (rank : 81) | θ value | 0.0193708 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.039542 (rank : 82) | θ value | 0.163984 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.038724 (rank : 83) | θ value | 0.0431538 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.038465 (rank : 84) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.038314 (rank : 85) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.038178 (rank : 86) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.037967 (rank : 87) | θ value | 0.0563607 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.035246 (rank : 88) | θ value | 0.0431538 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
TM108_HUMAN
|
||||||
| NC score | 0.033881 (rank : 89) | θ value | 0.163984 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.033154 (rank : 90) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.033043 (rank : 91) | θ value | 1.06291 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.032870 (rank : 92) | θ value | 0.813845 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.031812 (rank : 93) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.031798 (rank : 94) | θ value | 0.62314 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.030833 (rank : 95) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CO4A6_HUMAN
|
||||||
| NC score | 0.030322 (rank : 96) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14031, Q12823, Q14053, Q5JYH6, Q9NQM5, Q9NTX3, Q9UJ76, Q9UMG6, Q9Y4L4 | Gene names | COL4A6 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-6(IV) chain precursor. | |||||
|
GLDN_HUMAN
|
||||||
| NC score | 0.027853 (rank : 97) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZMI3, Q6UXZ7, Q7Z359 | Gene names | GLDN, COLM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gliomedin. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.026116 (rank : 98) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.024895 (rank : 99) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.024017 (rank : 100) | θ value | 0.62314 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.022771 (rank : 101) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.022396 (rank : 102) | θ value | 0.47712 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.022392 (rank : 103) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SGIP1_HUMAN
|
||||||
| NC score | 0.021379 (rank : 104) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BQI5, Q4LE32, Q5VYE2, Q5VYE3, Q5VYE4, Q68D76, Q6MZY6, Q8IWC2 | Gene names | SGIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.021255 (rank : 105) | θ value | 1.38821 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.020530 (rank : 106) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MO4L2_MOUSE
|
||||||
| NC score | 0.020213 (rank : 107) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
MO4L2_HUMAN
|
||||||
| NC score | 0.020092 (rank : 108) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.019426 (rank : 109) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.018948 (rank : 110) | θ value | 0.62314 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.018861 (rank : 111) | θ value | 0.47712 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.018364 (rank : 112) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.017236 (rank : 113) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TYDP1_MOUSE
|
||||||
| NC score | 0.016885 (rank : 114) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ37, Q8VE39 | Gene names | Tdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosyl-DNA phosphodiesterase 1 (EC 3.1.4.-) (Tyr-DNA phosphodiesterase 1). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.016653 (rank : 115) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.015283 (rank : 116) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.014503 (rank : 117) | θ value | 0.365318 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.014184 (rank : 118) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.014078 (rank : 119) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.012984 (rank : 120) | θ value | 0.21417 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
PACE1_MOUSE
|
||||||
| NC score | 0.012323 (rank : 121) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBQ7, Q6NY01, Q8BQC9, Q8BRJ1 | Gene names | Scyl3, Pace1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-associating with the carboxyl-terminal domain of ezrin (Ezrin- binding protein PACE-1) (SCY1-like protein 3). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.012236 (rank : 122) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
LAMP3_HUMAN
|
||||||
| NC score | 0.009884 (rank : 123) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQV4, O94781, Q8NEC8 | Gene names | LAMP3, DCLAMP, TSC403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosome-associated membrane glycoprotein 3 precursor (LAMP-3) (Lysosomal-associated membrane protein 3) (DC-lysosome-associated membrane glycoprotein) (DC LAMP) (Protein TSC403) (CD208 antigen). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.009479 (rank : 124) | θ value | 1.38821 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
PMS2_MOUSE
|
||||||
| NC score | 0.009388 (rank : 125) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.007928 (rank : 126) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
DGKG_MOUSE
|
||||||
| NC score | 0.007712 (rank : 127) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.007641 (rank : 128) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.006987 (rank : 129) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
LAEVR_HUMAN
|
||||||
| NC score | 0.006953 (rank : 130) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6Q4G3, Q32MR1, Q4G0I9, Q4G0V2, Q86XA3, Q8NBZ2 | Gene names | LVRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laeverin (EC 3.4.-.-) (CHL2 antigen). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.006561 (rank : 131) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.006524 (rank : 132) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.006345 (rank : 133) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.006229 (rank : 134) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
KPBB_HUMAN
|
||||||
| NC score | 0.006156 (rank : 135) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93100, Q8N4T5 | Gene names | PHKB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit beta (Phosphorylase kinase subunit beta). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.005984 (rank : 136) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.004950 (rank : 137) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CO2_HUMAN
|
||||||
| NC score | 0.004937 (rank : 138) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
PDIA5_MOUSE
|
||||||
| NC score | 0.004428 (rank : 139) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921X9 | Gene names | Pdia5, Pdir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase A5 precursor (EC 5.3.4.1) (Protein disulfide isomerase-related protein). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.004169 (rank : 140) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.003597 (rank : 141) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
FOXK1_MOUSE
|
||||||
| NC score | 0.003545 (rank : 142) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.002815 (rank : 143) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.002597 (rank : 144) | θ value | 0.0431538 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.002491 (rank : 145) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.002472 (rank : 146) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.001143 (rank : 147) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | -0.001288 (rank : 148) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
EPHB3_MOUSE
|
||||||
| NC score | -0.002445 (rank : 149) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54754, Q62214 | Gene names | Ephb3, Etk2, Mdk5, Sek4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-5) (Developmental kinase 5) (SEK-4). | |||||
|
EPHB3_HUMAN
|
||||||
| NC score | -0.002457 (rank : 150) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54753, Q7Z740 | Gene names | EPHB3, ETK2, HEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HEK-2). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | -0.002853 (rank : 151) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||