Please be patient as the page loads
|
MO4L2_MOUSE
|
||||||
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MO4L2_MOUSE
|
||||||
| θ value | 8.86892e-165 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
MO4L2_HUMAN
|
||||||
| θ value | 6.1562e-158 (rank : 2) | NC score | 0.985386 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
MO4L1_HUMAN
|
||||||
| θ value | 1.13764e-111 (rank : 3) | NC score | 0.959248 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBU8, O95899, Q5QTS1, Q6NVX8, Q86YT7, Q9HBP6, Q9NSW5 | Gene names | MORF4L1, MRG15 | |||
|
Domain Architecture |
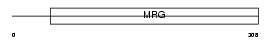
|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (MSL3-1 protein). | |||||
|
MO4L1_MOUSE
|
||||||
| θ value | 1.13764e-111 (rank : 4) | NC score | 0.958993 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60762, Q8BNY9, Q99MQ0 | Gene names | Morf4l1, Mrg15, Tex189 | |||
|
Domain Architecture |
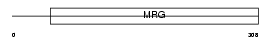
|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (Testis-expressed gene 189 protein). | |||||
|
MORF4_HUMAN
|
||||||
| θ value | 1.94503e-103 (rank : 5) | NC score | 0.973919 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y690 | Gene names | MORF4 | |||
|
Domain Architecture |
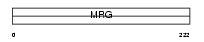
|
|||||
| Description | Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence-related protein 1) (SEN1). | |||||
|
MS3L1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 6) | NC score | 0.448705 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 7) | NC score | 0.430745 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 8) | NC score | 0.073552 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
REXO1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.086875 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
H15_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 10) | NC score | 0.081855 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
WFS1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.075391 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O76024, Q8N6I3, Q9UNW6 | Gene names | WFS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wolframin. | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.020821 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
H15_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.076872 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.026286 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LMX1B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.026030 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60663, O75463 | Gene names | LMX1B | |||
|
Domain Architecture |
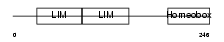
|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
LMX1B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.025877 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88609 | Gene names | Lmx1b | |||
|
Domain Architecture |
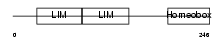
|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.032078 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.028613 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
MRP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.089986 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.067853 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.021058 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
H14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.066420 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.030273 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
NOL3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.037024 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |
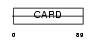
|
|||||
| Description | Nucleolar protein 3. | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.036589 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.027106 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.015960 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.028808 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.038900 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.029175 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.033906 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
H14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.071189 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
ING5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.027890 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D8Y8, Q3UL57, Q6P292, Q9CV64, Q9D9V8 | Gene names | Ing5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5. | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.067155 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MEOX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.011040 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50222, O75263, Q9UPL6 | Gene names | MEOX2, GAX, MOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2) (Growth arrest-specific homeobox). | |||||
|
SAKS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.025414 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
SORC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.020631 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.041328 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYT3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.007700 (rank : 87) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYTC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.035826 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0R2, Q3TL42, Q7TMQ2, Q8BMI6, Q9CX03 | Gene names | Tars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.020213 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.027489 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.010395 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.026926 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.039546 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.015991 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
FUS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.024140 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.066582 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
H13_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.062891 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
MECP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.030661 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2D6 | Gene names | Mecp2 | |||
|
Domain Architecture |
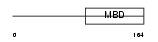
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
MEOX2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.010226 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P32443, Q544T6 | Gene names | Meox2, Gax, Mox-2, Mox2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.020458 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.040032 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.013761 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
IF5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.071425 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59325 | Gene names | Eif5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
MECP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.029181 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |
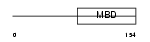
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
RCC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.014564 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
SOX3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.006529 (rank : 88) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.016292 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.016871 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.015185 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.026180 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
H12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.058767 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |
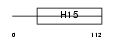
|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.016817 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
RCC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.013906 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
RHG26_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.008255 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.070923 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
ANX13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.004016 (rank : 89) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27216, Q9BQR5 | Gene names | ANXA13, ANX13 | |||
|
Domain Architecture |
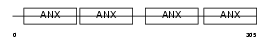
|
|||||
| Description | Annexin A13 (Annexin XIII) (Annexin, intestine-specific) (ISA). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.017937 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
CA104_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.018146 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66K80, Q8NA07 | Gene names | C1orf104 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf104. | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.009468 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CLAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.009287 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003678 (rank : 90) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.018815 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
H12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.057556 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |
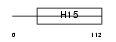
|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
IF2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.038713 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20042, Q9BVU0, Q9UJE4 | Gene names | EIF2S2, EIF2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta). | |||||
|
IPP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.027407 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERT9 | Gene names | Ppp1r1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase inhibitor 1 (IPP-1) (I-1). | |||||
|
NSMA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.010550 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.008657 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.012238 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.013294 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.010524 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.015660 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.033762 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.008397 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.013238 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
H11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050808 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |
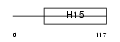
|
|||||
| Description | Histone H1.1. | |||||
|
H13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.052160 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |
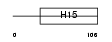
|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
IF5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051009 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55010, Q9H5N2, Q9UG48 | Gene names | EIF5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.072113 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
MO4L2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.86892e-165 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
MO4L2_HUMAN
|
||||||
| NC score | 0.985386 (rank : 2) | θ value | 6.1562e-158 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
MORF4_HUMAN
|
||||||
| NC score | 0.973919 (rank : 3) | θ value | 1.94503e-103 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y690 | Gene names | MORF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence-related protein 1) (SEN1). | |||||
|
MO4L1_HUMAN
|
||||||
| NC score | 0.959248 (rank : 4) | θ value | 1.13764e-111 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBU8, O95899, Q5QTS1, Q6NVX8, Q86YT7, Q9HBP6, Q9NSW5 | Gene names | MORF4L1, MRG15 | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (MSL3-1 protein). | |||||
|
MO4L1_MOUSE
|
||||||
| NC score | 0.958993 (rank : 5) | θ value | 1.13764e-111 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60762, Q8BNY9, Q99MQ0 | Gene names | Morf4l1, Mrg15, Tex189 | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 1 (MORF-related gene 15 protein) (Transcription factor-like protein MRG15) (Testis-expressed gene 189 protein). | |||||
|
MS3L1_HUMAN
|
||||||
| NC score | 0.448705 (rank : 6) | θ value | 1.29631e-06 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.430745 (rank : 7) | θ value | 1.69304e-06 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.089986 (rank : 8) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
REXO1_MOUSE
|
||||||
| NC score | 0.086875 (rank : 9) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TT28, Q3UMP1, Q69ZR0, Q6NSQ6, Q6PI95, Q9DA29 | Gene names | Rexo1, Kiaa1138, Tceb3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
H15_HUMAN
|
||||||
| NC score | 0.081855 (rank : 10) | θ value | 0.0148317 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
H15_MOUSE
|
||||||
| NC score | 0.076872 (rank : 11) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
WFS1_HUMAN
|
||||||
| NC score | 0.075391 (rank : 12) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O76024, Q8N6I3, Q9UNW6 | Gene names | WFS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wolframin. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.073552 (rank : 13) | θ value | 0.00134147 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.072113 (rank : 14) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
IF5_MOUSE
|
||||||
| NC score | 0.071425 (rank : 15) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59325 | Gene names | Eif5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.071189 (rank : 16) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.070923 (rank : 17) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.067853 (rank : 18) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.067155 (rank : 19) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.066582 (rank : 20) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.066420 (rank : 21) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.062891 (rank : 22) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
H12_MOUSE
|
||||||
| NC score | 0.058767 (rank : 23) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
H12_HUMAN
|
||||||
| NC score | 0.057556 (rank : 24) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
H13_HUMAN
|
||||||
| NC score | 0.052160 (rank : 25) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
IF5_HUMAN
|
||||||
| NC score | 0.051009 (rank : 26) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55010, Q9H5N2, Q9UG48 | Gene names | EIF5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5 (eIF-5). | |||||
|
H11_HUMAN
|
||||||
| NC score | 0.050808 (rank : 27) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.041328 (rank : 28) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.040032 (rank : 29) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.039546 (rank : 30) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.038900 (rank : 31) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
IF2B_HUMAN
|
||||||
| NC score | 0.038713 (rank : 32) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20042, Q9BVU0, Q9UJE4 | Gene names | EIF2S2, EIF2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2 subunit 2 (Eukaryotic translation initiation factor 2 subunit beta) (eIF-2-beta). | |||||
|
NOL3_MOUSE
|
||||||
| NC score | 0.037024 (rank : 33) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein 3. | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.036589 (rank : 34) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SYTC_MOUSE
|
||||||
| NC score | 0.035826 (rank : 35) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D0R2, Q3TL42, Q7TMQ2, Q8BMI6, Q9CX03 | Gene names | Tars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Threonyl-tRNA synthetase, cytoplasmic (EC 6.1.1.3) (Threonine--tRNA ligase) (ThrRS). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.033906 (rank : 36) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.033762 (rank : 37) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.032078 (rank : 38) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MECP2_MOUSE
|
||||||
| NC score | 0.030661 (rank : 39) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2D6 | Gene names | Mecp2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.030273 (rank : 40) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
MECP2_HUMAN
|
||||||
| NC score | 0.029181 (rank : 41) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.029175 (rank : 42) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.028808 (rank : 43) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.028613 (rank : 44) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
ING5_MOUSE
|
||||||
| NC score | 0.027890 (rank : 45) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D8Y8, Q3UL57, Q6P292, Q9CV64, Q9D9V8 | Gene names | Ing5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5. | |||||
|
UBP5_HUMAN
|
||||||
| NC score | 0.027489 (rank : 46) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P45974, Q96J22 | Gene names | USP5, ISOT | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 5 (EC 3.1.2.15) (Ubiquitin thioesterase 5) (Ubiquitin-specific-processing protease 5) (Deubiquitinating enzyme 5) (Isopeptidase T). | |||||
|
IPP1_MOUSE
|
||||||
| NC score | 0.027407 (rank : 47) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERT9 | Gene names | Ppp1r1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase inhibitor 1 (IPP-1) (I-1). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.027106 (rank : 48) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.026926 (rank : 49) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.026286 (rank : 50) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.026180 (rank : 51) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
LMX1B_HUMAN
|
||||||
| NC score | 0.026030 (rank : 52) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60663, O75463 | Gene names | LMX1B | |||
|
Domain Architecture |

|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
LMX1B_MOUSE
|
||||||
| NC score | 0.025877 (rank : 53) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88609 | Gene names | Lmx1b | |||
|
Domain Architecture |

|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
SAKS1_HUMAN
|
||||||
| NC score | 0.025414 (rank : 54) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04323, Q9BV93, Q9BVV5 | Gene names | SAKS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAPK substrate protein 1 (UBA/UBX 33.3 kDa protein). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.024140 (rank : 55) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.021058 (rank : 56) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.020821 (rank : 57) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
SORC2_MOUSE
|
||||||
| NC score | 0.020631 (rank : 58) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPR5 | Gene names | Sorcs2 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS2 precursor. | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.020458 (rank : 59) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.020213 (rank : 60) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.018815 (rank : 61) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
CA104_HUMAN
|
||||||
| NC score | 0.018146 (rank : 62) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66K80, Q8NA07 | Gene names | C1orf104 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf104. | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.017937 (rank : 63) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.016871 (rank : 64) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.016817 (rank : 65) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.016292 (rank : 66) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.015991 (rank : 67) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.015960 (rank : 68) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.015660 (rank : 69) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.015185 (rank : 70) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
RCC2_HUMAN
|
||||||
| NC score | 0.014564 (rank : 71) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P258, Q8IVL9, Q9BSN6, Q9NPV8 | Gene names | RCC2, KIAA1470, TD60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2 (Telophase disk protein of 60 kDa) (RCC1-like protein TD- 60). | |||||
|
RCC2_MOUSE
|
||||||
| NC score | 0.013906 (rank : 72) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BK67, Q6ZPQ0 | Gene names | Rcc2, Kiaa1470 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RCC2. | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.013761 (rank : 73) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.013294 (rank : 74) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.013238 (rank : 75) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.012238 (rank : 76) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MEOX2_HUMAN
|
||||||
| NC score | 0.011040 (rank : 77) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50222, O75263, Q9UPL6 | Gene names | MEOX2, GAX, MOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2) (Growth arrest-specific homeobox). | |||||
|
NSMA2_MOUSE
|
||||||
| NC score | 0.010550 (rank : 78) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.010524 (rank : 79) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.010395 (rank : 80) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
MEOX2_MOUSE
|
||||||
| NC score | 0.010226 (rank : 81) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P32443, Q544T6 | Gene names | Meox2, Gax, Mox-2, Mox2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.009468 (rank : 82) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CLAP1_HUMAN
|
||||||
| NC score | 0.009287 (rank : 83) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.008657 (rank : 84) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.008397 (rank : 85) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
RHG26_HUMAN
|
||||||
| NC score | 0.008255 (rank : 86) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNA1, O75117, Q9BYS6, Q9BYS7, Q9UJ00 | Gene names | ARHGAP26, GRAF, KIAA0621, OPHN1L | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 26 (Oligophrenin-1-like protein) (GTPase regulator associated with focal adhesion kinase). | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.007700 (rank : 87) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SOX3_MOUSE
|
||||||
| NC score | 0.006529 (rank : 88) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53784 | Gene names | Sox3, Sox-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
ANX13_HUMAN
|
||||||
| NC score | 0.004016 (rank : 89) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27216, Q9BQR5 | Gene names | ANXA13, ANX13 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A13 (Annexin XIII) (Annexin, intestine-specific) (ISA). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.003678 (rank : 90) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||