Please be patient as the page loads
|
SYEP_MOUSE
|
||||||
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SYEP_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.965214 (rank : 2) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYQ_HUMAN
|
||||||
| θ value | 1.36269e-64 (rank : 3) | NC score | 0.774639 (rank : 3) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47897 | Gene names | QARS | |||
|
Domain Architecture |

|
|||||
| Description | Glutaminyl-tRNA synthetase (EC 6.1.1.18) (Glutamine--tRNA ligase) (GlnRS). | |||||
|
SYW_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 4) | NC score | 0.384037 (rank : 4) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P32921, Q80ZY4, Q99J58, Q9DC65 | Gene names | Wars, Wrs | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophanyl-tRNA synthetase (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS). | |||||
|
SYW_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 5) | NC score | 0.350932 (rank : 5) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23381, P78535, Q9UDL3 | Gene names | WARS, WRS | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophanyl-tRNA synthetase (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) (IFP53) (hWRS). | |||||
|
SYM_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 6) | NC score | 0.221719 (rank : 8) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
MCA3_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 7) | NC score | 0.268022 (rank : 6) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D1M4 | Gene names | Eef1e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
MCA3_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 8) | NC score | 0.247800 (rank : 7) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |
|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
SYM_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 9) | NC score | 0.199997 (rank : 9) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 10) | NC score | 0.118691 (rank : 13) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 11) | NC score | 0.059247 (rank : 43) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.083867 (rank : 23) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SYG_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 13) | NC score | 0.146568 (rank : 11) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41250, Q969Y1 | Gene names | GARS | |||
|
Domain Architecture |

|
|||||
| Description | Glycyl-tRNA synthetase (EC 6.1.1.14) (Glycine--tRNA ligase) (GlyRS). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.068441 (rank : 36) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 15) | NC score | 0.042356 (rank : 75) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
SYG_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 16) | NC score | 0.163563 (rank : 10) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CZD3, Q8VC67 | Gene names | Gars | |||
|
Domain Architecture |

|
|||||
| Description | Glycyl-tRNA synthetase (EC 6.1.1.14) (Glycine--tRNA ligase) (GlyRS). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 17) | NC score | 0.094865 (rank : 17) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 18) | NC score | 0.056565 (rank : 49) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CD2AP_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.057818 (rank : 45) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |
|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
RGS6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 20) | NC score | 0.046668 (rank : 70) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.082505 (rank : 24) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 22) | NC score | 0.030413 (rank : 94) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
SYV_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.079501 (rank : 26) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |
|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
H13_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.104530 (rank : 14) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.074032 (rank : 29) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
H14_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.100569 (rank : 15) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.052125 (rank : 60) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SYH_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.089349 (rank : 20) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12081 | Gene names | HARS, HRS | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS). | |||||
|
H14_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.099517 (rank : 16) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
MCA2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.139837 (rank : 12) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13155, Q96CZ5, Q9P1L2 | Gene names | JTV1 | |||
|
Domain Architecture |
|
|||||
| Description | Multisynthetase complex auxiliary component p38 (Protein JTV-1). | |||||
|
LETM1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.055132 (rank : 50) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.049089 (rank : 66) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.053665 (rank : 52) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.057712 (rank : 47) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
SYH_MOUSE
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.092030 (rank : 19) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61035 | Gene names | Hars | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.078371 (rank : 27) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.040405 (rank : 77) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.072519 (rank : 32) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.047577 (rank : 69) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.033575 (rank : 88) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CD2AP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.046441 (rank : 71) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5K6, Q9UG97 | Gene names | CD2AP | |||
|
Domain Architecture |
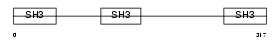
|
|||||
| Description | CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.064728 (rank : 37) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.040628 (rank : 76) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SYCC_MOUSE
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.052955 (rank : 54) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
H15_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.092711 (rank : 18) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
H15_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.089283 (rank : 21) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.080285 (rank : 25) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.070925 (rank : 33) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.059311 (rank : 42) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SYPM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.078317 (rank : 28) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFI5, Q8R0C4 | Gene names | Pars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable prolyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.15) (Proline--tRNA ligase) (ProRS). | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.015956 (rank : 123) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
H12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.084142 (rank : 22) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |
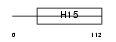
|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.021303 (rank : 114) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.034273 (rank : 85) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.045704 (rank : 72) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SCG2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.049356 (rank : 64) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03517, Q80Y79, Q9CW80 | Gene names | Scg2, Chgc, Scg-2 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
SYV_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.060946 (rank : 41) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.069385 (rank : 34) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.028581 (rank : 99) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.031381 (rank : 92) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MO4L2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.044731 (rank : 73) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
RGS6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.033664 (rank : 87) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.044298 (rank : 74) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.026928 (rank : 100) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.025342 (rank : 105) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.047756 (rank : 68) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NCF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.049030 (rank : 67) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19878, Q2PP06, Q8NFC7, Q9BV51 | Gene names | NCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 2 (NCF-2) (Neutrophil NADPH oxidase factor 2) (67 kDa neutrophil oxidase factor) (p67-phox) (NOXA2). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.033741 (rank : 86) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.050416 (rank : 63) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.031597 (rank : 91) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
MO4L2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.039546 (rank : 79) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.021623 (rank : 111) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
OST48_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.038819 (rank : 80) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39656, O43244, Q8NI93, Q9BUI2 | Gene names | DDOST, KIAA0115, OST48 | |||
|
Domain Architecture |
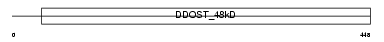
|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit precursor (EC 2.4.1.119) (Oligosaccharyl transferase 48 kDa subunit) (DDOST 48 kDa subunit). | |||||
|
OST48_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.038715 (rank : 81) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54734 | Gene names | Ddost | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit precursor (EC 2.4.1.119) (Oligosaccharyl transferase 48 kDa subunit) (DDOST 48 kDa subunit). | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.017350 (rank : 121) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.017433 (rank : 120) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.016710 (rank : 122) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.021514 (rank : 112) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
EF1G_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.073218 (rank : 30) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |
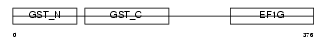
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.059068 (rank : 44) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
RGS7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.029874 (rank : 96) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
TLR4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.007317 (rank : 133) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUK6, Q9D691, Q9QZF5, Q9Z203 | Gene names | Tlr4, Lps | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 4 precursor (CD284 antigen). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.025294 (rank : 106) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ARS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.032455 (rank : 89) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
GP155_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.054705 (rank : 51) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z3F1, Q53SJ3, Q53TA8, Q69YG8, Q86SP9, Q8N261, Q8N639, Q8N8K3, Q96MV6 | Gene names | GPR155, PGR22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR155 (G-protein coupled receptor PGR22). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.061727 (rank : 40) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RAD52_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.024526 (rank : 107) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43351, Q13205 | Gene names | RAD52 | |||
|
Domain Architecture |
|
|||||
| Description | DNA repair protein RAD52 homolog. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.039701 (rank : 78) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.026013 (rank : 103) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.019437 (rank : 117) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MCES_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.036363 (rank : 83) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.049264 (rank : 65) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.017913 (rank : 119) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.022285 (rank : 110) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
EF1G_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.056802 (rank : 48) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
LETM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.038632 (rank : 82) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z2I0, Q5PQC5, Q7TMK8, Q80ZX6, Q8CGJ3, Q8K5E5 | Gene names | Letm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
MORF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.026649 (rank : 102) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y690 | Gene names | MORF4 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence-related protein 1) (SEN1). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.024105 (rank : 108) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.012926 (rank : 127) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.018695 (rank : 118) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SCG2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.032443 (rank : 90) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
SHE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.012028 (rank : 128) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BSD5, Q3TZT0 | Gene names | She | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.025351 (rank : 104) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.026769 (rank : 101) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.021237 (rank : 115) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
H12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.073041 (rank : 31) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
HOME2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.024092 (rank : 109) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NSB8, O95269, O95349, Q9NSB6, Q9NSB7, Q9UNT7 | Gene names | HOMER2 | |||
|
Domain Architecture |
|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.029055 (rank : 98) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.021474 (rank : 113) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
INADL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.007812 (rank : 132) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.020494 (rank : 116) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.034620 (rank : 84) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.014290 (rank : 126) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PTN6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.009472 (rank : 131) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.029411 (rank : 97) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.006174 (rank : 134) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
TLN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.010845 (rank : 130) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.010908 (rank : 129) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.029953 (rank : 95) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.014355 (rank : 124) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.014315 (rank : 125) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.030482 (rank : 93) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
ZN583_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | -0.002507 (rank : 135) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96ND8, O14850, Q2NKK3 | Gene names | ZNF583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583 (Zinc finger protein L3-5). | |||||
|
H10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.053545 (rank : 53) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
H10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.052387 (rank : 58) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
H11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.061928 (rank : 39) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1. | |||||
|
H11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.064336 (rank : 38) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
H13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.069362 (rank : 35) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
H1T_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.051285 (rank : 62) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22492, Q8IUE8 | Gene names | HIST1H1T, H1FT, H1T | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
H1T_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052710 (rank : 55) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07133, Q8CGP8 | Gene names | Hist1h1t, H1ft, H1t | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.051395 (rank : 61) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MINT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052249 (rank : 59) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052406 (rank : 57) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SYPM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.052678 (rank : 56) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L3T8, Q9H6S5, Q9UFT1 | Gene names | PARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable prolyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.15) (Proline--tRNA ligase) (ProRS). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.057792 (rank : 46) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYEP_HUMAN
|
||||||
| NC score | 0.965214 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYQ_HUMAN
|
||||||
| NC score | 0.774639 (rank : 3) | θ value | 1.36269e-64 (rank : 3) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47897 | Gene names | QARS | |||
|
Domain Architecture |

|
|||||
| Description | Glutaminyl-tRNA synthetase (EC 6.1.1.18) (Glutamine--tRNA ligase) (GlnRS). | |||||
|
SYW_MOUSE
|
||||||
| NC score | 0.384037 (rank : 4) | θ value | 2.98157e-11 (rank : 4) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P32921, Q80ZY4, Q99J58, Q9DC65 | Gene names | Wars, Wrs | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophanyl-tRNA synthetase (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS). | |||||
|
SYW_HUMAN
|
||||||
| NC score | 0.350932 (rank : 5) | θ value | 1.06045e-08 (rank : 5) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23381, P78535, Q9UDL3 | Gene names | WARS, WRS | |||
|
Domain Architecture |

|
|||||
| Description | Tryptophanyl-tRNA synthetase (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) (IFP53) (hWRS). | |||||
|
MCA3_MOUSE
|
||||||
| NC score | 0.268022 (rank : 6) | θ value | 1.87187e-05 (rank : 7) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D1M4 | Gene names | Eef1e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
MCA3_HUMAN
|
||||||
| NC score | 0.247800 (rank : 7) | θ value | 4.1701e-05 (rank : 8) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
SYM_HUMAN
|
||||||
| NC score | 0.221719 (rank : 8) | θ value | 1.09739e-05 (rank : 6) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
SYM_MOUSE
|
||||||
| NC score | 0.199997 (rank : 9) | θ value | 9.29e-05 (rank : 9) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
SYG_MOUSE
|
||||||
| NC score | 0.163563 (rank : 10) | θ value | 0.00665767 (rank : 16) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CZD3, Q8VC67 | Gene names | Gars | |||
|
Domain Architecture |

|
|||||
| Description | Glycyl-tRNA synthetase (EC 6.1.1.14) (Glycine--tRNA ligase) (GlyRS). | |||||
|
SYG_HUMAN
|
||||||
| NC score | 0.146568 (rank : 11) | θ value | 0.00390308 (rank : 13) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41250, Q969Y1 | Gene names | GARS | |||
|
Domain Architecture |

|
|||||
| Description | Glycyl-tRNA synthetase (EC 6.1.1.14) (Glycine--tRNA ligase) (GlyRS). | |||||
|
MCA2_HUMAN
|
||||||
| NC score | 0.139837 (rank : 12) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13155, Q96CZ5, Q9P1L2 | Gene names | JTV1 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p38 (Protein JTV-1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.118691 (rank : 13) | θ value | 0.000786445 (rank : 10) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.104530 (rank : 14) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.100569 (rank : 15) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.099517 (rank : 16) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.094865 (rank : 17) | θ value | 0.00665767 (rank : 17) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
H15_HUMAN
|
||||||
| NC score | 0.092711 (rank : 18) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
SYH_MOUSE
|
||||||
| NC score | 0.092030 (rank : 19) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61035 | Gene names | Hars | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS). | |||||
|
SYH_HUMAN
|
||||||
| NC score | 0.089349 (rank : 20) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12081 | Gene names | HARS, HRS | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase (EC 6.1.1.21) (Histidine--tRNA ligase) (HisRS). | |||||
|
H15_MOUSE
|
||||||
| NC score | 0.089283 (rank : 21) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
H12_HUMAN
|
||||||
| NC score | 0.084142 (rank : 22) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.083867 (rank : 23) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.082505 (rank : 24) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.080285 (rank : 25) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SYV_MOUSE
|
||||||
| NC score | 0.079501 (rank : 26) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.078371 (rank : 27) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SYPM_MOUSE
|
||||||
| NC score | 0.078317 (rank : 28) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFI5, Q8R0C4 | Gene names | Pars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable prolyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.15) (Proline--tRNA ligase) (ProRS). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.074032 (rank : 29) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
EF1G_HUMAN
|
||||||
| NC score | 0.073218 (rank : 30) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
H12_MOUSE
|
||||||
| NC score | 0.073041 (rank : 31) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.072519 (rank : 32) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.070925 (rank : 33) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.069385 (rank : 34) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
H13_HUMAN
|
||||||
| NC score | 0.069362 (rank : 35) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.068441 (rank : 36) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.064728 (rank : 37) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
H11_MOUSE
|
||||||
| NC score | 0.064336 (rank : 38) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
H11_HUMAN
|
||||||
| NC score | 0.061928 (rank : 39) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.061727 (rank : 40) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SYV_HUMAN
|
||||||
| NC score | 0.060946 (rank : 41) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.059311 (rank : 42) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.059247 (rank : 43) | θ value | 0.00175202 (rank : 11) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.059068 (rank : 44) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
CD2AP_MOUSE
|
||||||
| NC score | 0.057818 (rank : 45) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.057792 (rank : 46) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.057712 (rank : 47) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
EF1G_MOUSE
|
||||||
| NC score | 0.056802 (rank : 48) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.056565 (rank : 49) | θ value | 0.0148317 (rank : 18) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LETM1_HUMAN
|
||||||
| NC score | 0.055132 (rank : 50) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95202, Q9UF65 | Gene names | LETM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
GP155_HUMAN
|
||||||
| NC score | 0.054705 (rank : 51) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z3F1, Q53SJ3, Q53TA8, Q69YG8, Q86SP9, Q8N261, Q8N639, Q8N8K3, Q96MV6 | Gene names | GPR155, PGR22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR155 (G-protein coupled receptor PGR22). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.053665 (rank : 52) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
H10_HUMAN
|
||||||
| NC score | 0.053545 (rank : 53) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
SYCC_MOUSE
|
||||||
| NC score | 0.052955 (rank : 54) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
H1T_MOUSE
|
||||||
| NC score | 0.052710 (rank : 55) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q07133, Q8CGP8 | Gene names | Hist1h1t, H1ft, H1t | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
SYPM_HUMAN
|
||||||
| NC score | 0.052678 (rank : 56) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L3T8, Q9H6S5, Q9UFT1 | Gene names | PARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable prolyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.15) (Proline--tRNA ligase) (ProRS). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.052406 (rank : 57) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
H10_MOUSE
|
||||||
| NC score | 0.052387 (rank : 58) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.052249 (rank : 59) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.052125 (rank : 60) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.051395 (rank : 61) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
H1T_HUMAN
|
||||||
| NC score | 0.051285 (rank : 62) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22492, Q8IUE8 | Gene names | HIST1H1T, H1FT, H1T | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.050416 (rank : 63) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
SCG2_MOUSE
|
||||||
| NC score | 0.049356 (rank : 64) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03517, Q80Y79, Q9CW80 | Gene names | Scg2, Chgc, Scg-2 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.049264 (rank : 65) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.049089 (rank : 66) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NCF2_HUMAN
|
||||||
| NC score | 0.049030 (rank : 67) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19878, Q2PP06, Q8NFC7, Q9BV51 | Gene names | NCF2 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 2 (NCF-2) (Neutrophil NADPH oxidase factor 2) (67 kDa neutrophil oxidase factor) (p67-phox) (NOXA2). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.047756 (rank : 68) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.047577 (rank : 69) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
RGS6_HUMAN
|
||||||
| NC score | 0.046668 (rank : 70) | θ value | 0.0193708 (rank : 20) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
CD2AP_HUMAN
|
||||||
| NC score | 0.046441 (rank : 71) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5K6, Q9UG97 | Gene names | CD2AP | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Cas ligand with multiple SH3 domains) (Adapter protein CMS). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.045704 (rank : 72) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MO4L2_HUMAN
|
||||||
| NC score | 0.044731 (rank : 73) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15014, Q567V0, Q8J026 | Gene names | MORF4L2, KIAA0026, MRGX | |||
|
Domain Architecture |

|
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (MSL3-2 protein). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.044298 (rank : 74) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.042356 (rank : 75) | θ value | 0.00509761 (rank : 15) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.040628 (rank : 76) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.040405 (rank : 77) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.039701 (rank : 78) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MO4L2_MOUSE
|
||||||
| NC score | 0.039546 (rank : 79) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0Q4, Q3UL62, Q6DIB1, Q6ZQK7, Q8C201, Q8C6C2 | Gene names | Morf4l2, Kiaa0026, Sid393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mortality factor 4-like protein 2 (MORF-related gene X protein) (Transcription factor-like protein MRGX) (Sid 393). | |||||
|
OST48_HUMAN
|
||||||
| NC score | 0.038819 (rank : 80) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39656, O43244, Q8NI93, Q9BUI2 | Gene names | DDOST, KIAA0115, OST48 | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit precursor (EC 2.4.1.119) (Oligosaccharyl transferase 48 kDa subunit) (DDOST 48 kDa subunit). | |||||
|
OST48_MOUSE
|
||||||
| NC score | 0.038715 (rank : 81) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54734 | Gene names | Ddost | |||
|
Domain Architecture |

|
|||||
| Description | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit precursor (EC 2.4.1.119) (Oligosaccharyl transferase 48 kDa subunit) (DDOST 48 kDa subunit). | |||||
|
LETM1_MOUSE
|
||||||
| NC score | 0.038632 (rank : 82) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z2I0, Q5PQC5, Q7TMK8, Q80ZX6, Q8CGJ3, Q8K5E5 | Gene names | Letm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
MCES_MOUSE
|
||||||
| NC score | 0.036363 (rank : 83) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.034620 (rank : 84) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.034273 (rank : 85) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.033741 (rank : 86) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
RGS6_MOUSE
|
||||||
| NC score | 0.033664 (rank : 87) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.033575 (rank : 88) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.032455 (rank : 89) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
SCG2_HUMAN
|
||||||
| NC score | 0.032443 (rank : 90) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.031597 (rank : 91) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.031381 (rank : 92) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.030482 (rank : 93) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.030413 (rank : 94) | θ value | 0.0252991 (rank : 22) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.029953 (rank : 95) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
RGS7_MOUSE
|
||||||
| NC score | 0.029874 (rank : 96) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.029411 (rank : 97) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.029055 (rank : 98) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.028581 (rank : 99) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.026928 (rank : 100) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.026769 (rank : 101) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MORF4_HUMAN
|
||||||
| NC score | 0.026649 (rank : 102) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y690 | Gene names | MORF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence-related protein 1) (SEN1). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.026013 (rank : 103) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.025351 (rank : 104) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.025342 (rank : 105) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.025294 (rank : 106) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
RAD52_HUMAN
|
||||||
| NC score | 0.024526 (rank : 107) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43351, Q13205 | Gene names | RAD52 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair protein RAD52 homolog. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.024105 (rank : 108) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
HOME2_HUMAN
|
||||||
| NC score | 0.024092 (rank : 109) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NSB8, O95269, O95349, Q9NSB6, Q9NSB7, Q9UNT7 | Gene names | HOMER2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin). | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.022285 (rank : 110) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.021623 (rank : 111) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.021514 (rank : 112) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.021474 (rank : 113) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.021303 (rank : 114) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.021237 (rank : 115) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.020494 (rank : 116) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.019437 (rank : 117) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.018695 (rank : 118) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.017913 (rank : 119) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.017433 (rank : 120) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.017350 (rank : 121) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.016710 (rank : 122) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.015956 (rank : 123) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.014355 (rank : 124) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.014315 (rank : 125) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.014290 (rank : 126) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.012926 (rank : 127) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
SHE_MOUSE
|
||||||
| NC score | 0.012028 (rank : 128) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BSD5, Q3TZT0 | Gene names | She | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
TLN1_MOUSE
|
||||||
| NC score | 0.010908 (rank : 129) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26039, Q8VEF0 | Gene names | Tln1, Tln | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
TLN1_HUMAN
|
||||||
| NC score | 0.010845 (rank : 130) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y490, Q86YD0, Q9NZQ2, Q9UHH8, Q9UPX3 | Gene names | TLN1, KIAA1027, TLN | |||
|
Domain Architecture |

|
|||||
| Description | Talin-1. | |||||
|
PTN6_HUMAN
|
||||||
| NC score | 0.009472 (rank : 131) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29350, Q969V8 | Gene names | PTPN6, HCP, PTP1C | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 6 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1C) (PTP-1C) (Hematopoietic cell protein-tyrosine phosphatase) (SH-PTP1) (Protein-tyrosine phosphatase SHP-1). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.007812 (rank : 132) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
TLR4_MOUSE
|
||||||
| NC score | 0.007317 (rank : 133) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUK6, Q9D691, Q9QZF5, Q9Z203 | Gene names | Tlr4, Lps | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 4 precursor (CD284 antigen). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.006174 (rank : 134) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ZN583_HUMAN
|
||||||
| NC score | -0.002507 (rank : 135) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96ND8, O14850, Q2NKK3 | Gene names | ZNF583 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 583 (Zinc finger protein L3-5). | |||||