Please be patient as the page loads
|
EF1G_HUMAN
|
||||||
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |
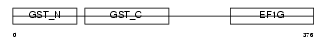
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EF1G_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
EF1G_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999017 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |
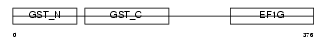
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
SYV_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 3) | NC score | 0.560372 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYV_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 4) | NC score | 0.552033 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |
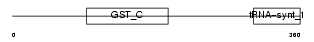
|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
MCA3_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 5) | NC score | 0.339836 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D1M4 | Gene names | Eef1e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
GSTT2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 6) | NC score | 0.185368 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61133, Q61134, Q64472 | Gene names | Gstt2 | |||
|
Domain Architecture |
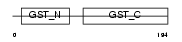
|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
GSTT2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 7) | NC score | 0.197731 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30712, O60665, Q9HD76 | Gene names | GSTT2 | |||
|
Domain Architecture |
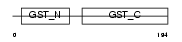
|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
MCA3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.259930 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |
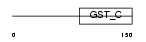
|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
PTGD2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.132547 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60760 | Gene names | PTGDS2, PGDS | |||
|
Domain Architecture |
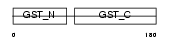
|
|||||
| Description | Glutathione-requiring prostaglandin D synthase (EC 5.3.99.2) (Glutathione-dependent PGD synthetase) (Prostaglandin-H2 D-isomerase) (Hematopoietic prostaglandin D synthase) (H-PGDS). | |||||
|
NCOAT_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.137632 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
NCOAT_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.137852 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
PTGD2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.118636 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JHF7 | Gene names | Ptgds2, Pgds | |||
|
Domain Architecture |
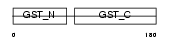
|
|||||
| Description | Glutathione-requiring prostaglandin D synthase (EC 5.3.99.2) (Glutathione-dependent PGD synthetase) (Prostaglandin-H2 D-isomerase) (Hematopoietic prostaglandin D synthase) (H-PGDS). | |||||
|
HA1B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.019327 (rank : 36) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01901 | Gene names | H2-K1, H2-K | |||
|
Domain Architecture |
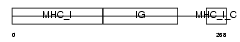
|
|||||
| Description | H-2 class I histocompatibility antigen, K-B alpha chain precursor (H- 2K(B)). | |||||
|
SYM_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.102626 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
GSTA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.059156 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15217, Q5T7Q8, Q9BX18, Q9H414 | Gene names | GSTA4 | |||
|
Domain Architecture |
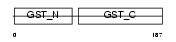
|
|||||
| Description | Glutathione S-transferase A4 (EC 2.5.1.18) (Glutathione S-transferase A4-4) (GST class-alpha member 4). | |||||
|
GSTT1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.123067 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64471 | Gene names | Gstt1 | |||
|
Domain Architecture |
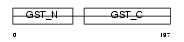
|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1). | |||||
|
HA1D_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.017170 (rank : 37) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01902 | Gene names | H2-K1, H2-K | |||
|
Domain Architecture |
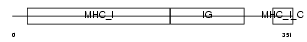
|
|||||
| Description | H-2 class I histocompatibility antigen, K-D alpha chain precursor (H- 2K(D)). | |||||
|
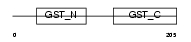
GSTO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.040578 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78417, Q5TA03, Q7Z3T2 | Gene names | GSTO1, GSTTLP28 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.046901 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.036176 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SIA7A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.032451 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SYM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.072645 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.073218 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYEP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.048473 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
B3GL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.020608 (rank : 35) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920V1, O54906, Q91V72, Q920V0, Q9CTE5 | Gene names | b3galnt1, B3galt3, B3gt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
GSTP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.041258 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09211, O00460, Q15690 | Gene names | GSTP1, GST3 | |||
|
Domain Architecture |
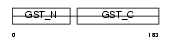
|
|||||
| Description | Glutathione S-transferase P (EC 2.5.1.18) (GST class-pi) (GSTP1-1). | |||||
|
HA1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.011636 (rank : 39) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01897, Q31195, Q31196, Q31197 | Gene names | H2-L | |||
|
Domain Architecture |
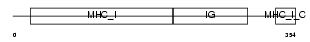
|
|||||
| Description | H-2 class I histocompatibility antigen, L-D alpha chain precursor. | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.021697 (rank : 34) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SYCC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.026065 (rank : 33) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.015956 (rank : 38) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
GSTT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.081120 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30711, O00226, Q969K8, Q96IY3 | Gene names | GSTT1 | |||
|
Domain Architecture |
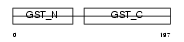
|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1) (Glutathione transferase T1-1). | |||||
|
SYIC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.118829 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41252, Q5TCD0, Q9H588 | Gene names | IARS | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
SYIC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.115802 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
SYIM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.141125 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NSE4, Q6PI85, Q7L439, Q86WU9, Q96D91 | Gene names | IARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.142457 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BIJ6, Q3TKT8, Q8BIJ0, Q8BIP3 | Gene names | Iars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYLM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.087338 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.082671 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDC0 | Gene names | Lars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYMM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.056772 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96GW9, Q76E79, Q8IW62, Q8N7N4 | Gene names | MARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYMM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.065027 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
EF1G_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
EF1G_MOUSE
|
||||||
| NC score | 0.999017 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
SYV_HUMAN
|
||||||
| NC score | 0.560372 (rank : 3) | θ value | 7.80994e-28 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26640, Q5JQ90, Q96E77, Q9UQM2 | Gene names | VARS, G7A, VARS2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
SYV_MOUSE
|
||||||
| NC score | 0.552033 (rank : 4) | θ value | 5.59698e-26 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
MCA3_MOUSE
|
||||||
| NC score | 0.339836 (rank : 5) | θ value | 1.43324e-05 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D1M4 | Gene names | Eef1e1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
MCA3_HUMAN
|
||||||
| NC score | 0.259930 (rank : 6) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
GSTT2_HUMAN
|
||||||
| NC score | 0.197731 (rank : 7) | θ value | 0.00298849 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30712, O60665, Q9HD76 | Gene names | GSTT2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
GSTT2_MOUSE
|
||||||
| NC score | 0.185368 (rank : 8) | θ value | 0.00228821 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61133, Q61134, Q64472 | Gene names | Gstt2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
SYIM_MOUSE
|
||||||
| NC score | 0.142457 (rank : 9) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BIJ6, Q3TKT8, Q8BIJ0, Q8BIP3 | Gene names | Iars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
SYIM_HUMAN
|
||||||
| NC score | 0.141125 (rank : 10) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NSE4, Q6PI85, Q7L439, Q86WU9, Q96D91 | Gene names | IARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Isoleucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS). | |||||
|
NCOAT_MOUSE
|
||||||
| NC score | 0.137852 (rank : 11) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQQ9, Q3ULY7, Q6ZQ71, Q8BK05, Q8BTT2, Q8CFX2, Q9CSJ4, Q9CUR7 | Gene names | Mgea5, Hexc, Kiaa0679 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
NCOAT_HUMAN
|
||||||
| NC score | 0.137632 (rank : 12) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
PTGD2_HUMAN
|
||||||
| NC score | 0.132547 (rank : 13) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60760 | Gene names | PTGDS2, PGDS | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione-requiring prostaglandin D synthase (EC 5.3.99.2) (Glutathione-dependent PGD synthetase) (Prostaglandin-H2 D-isomerase) (Hematopoietic prostaglandin D synthase) (H-PGDS). | |||||
|
GSTT1_MOUSE
|
||||||
| NC score | 0.123067 (rank : 14) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64471 | Gene names | Gstt1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1). | |||||
|
SYIC_HUMAN
|
||||||
| NC score | 0.118829 (rank : 15) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P41252, Q5TCD0, Q9H588 | Gene names | IARS | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
PTGD2_MOUSE
|
||||||
| NC score | 0.118636 (rank : 16) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JHF7 | Gene names | Ptgds2, Pgds | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione-requiring prostaglandin D synthase (EC 5.3.99.2) (Glutathione-dependent PGD synthetase) (Prostaglandin-H2 D-isomerase) (Hematopoietic prostaglandin D synthase) (H-PGDS). | |||||
|
SYIC_MOUSE
|
||||||
| NC score | 0.115802 (rank : 17) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BU30 | Gene names | Iars | |||
|
Domain Architecture |

|
|||||
| Description | Isoleucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.5) (Isoleucine--tRNA ligase) (IleRS) (IRS). | |||||
|
SYM_HUMAN
|
||||||
| NC score | 0.102626 (rank : 18) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
SYLM_HUMAN
|
||||||
| NC score | 0.087338 (rank : 19) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLM_MOUSE
|
||||||
| NC score | 0.082671 (rank : 20) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDC0 | Gene names | Lars2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
GSTT1_HUMAN
|
||||||
| NC score | 0.081120 (rank : 21) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30711, O00226, Q969K8, Q96IY3 | Gene names | GSTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1) (Glutathione transferase T1-1). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.073218 (rank : 22) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYM_MOUSE
|
||||||
| NC score | 0.072645 (rank : 23) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
SYMM_MOUSE
|
||||||
| NC score | 0.065027 (rank : 24) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
GSTA4_HUMAN
|
||||||
| NC score | 0.059156 (rank : 25) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15217, Q5T7Q8, Q9BX18, Q9H414 | Gene names | GSTA4 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase A4 (EC 2.5.1.18) (Glutathione S-transferase A4-4) (GST class-alpha member 4). | |||||
|
SYMM_HUMAN
|
||||||
| NC score | 0.056772 (rank : 26) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96GW9, Q76E79, Q8IW62, Q8N7N4 | Gene names | MARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
SYEP_HUMAN
|
||||||
| NC score | 0.048473 (rank : 27) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.046901 (rank : 28) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
GSTP1_HUMAN
|
||||||
| NC score | 0.041258 (rank : 29) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09211, O00460, Q15690 | Gene names | GSTP1, GST3 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase P (EC 2.5.1.18) (GST class-pi) (GSTP1-1). | |||||
|
GSTO1_HUMAN
|
||||||
| NC score | 0.040578 (rank : 30) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78417, Q5TA03, Q7Z3T2 | Gene names | GSTO1, GSTTLP28 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.036176 (rank : 31) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SIA7A_MOUSE
|
||||||
| NC score | 0.032451 (rank : 32) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SYCC_MOUSE
|
||||||
| NC score | 0.026065 (rank : 33) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.021697 (rank : 34) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
B3GL1_MOUSE
|
||||||
| NC score | 0.020608 (rank : 35) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920V1, O54906, Q91V72, Q920V0, Q9CTE5 | Gene names | b3galnt1, B3galt3, B3gt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 (EC 2.4.1.79) (Beta-3-GalNAc-T1) (Beta-1,3-galactosyltransferase 3) (Beta-1,3- GalTase 3) (Beta3Gal-T3) (b3Gal-T3) (Galactosylgalactosylglucosylceramide beta-D-acetyl- galactosaminyltransferase) (UDP-N- acetylgalactosamine:globotriaosylceramide beta-1,3-N- acetylgalactosaminyltransferase) (Globoside synthase) (Beta-3-Gx-T3). | |||||
|
HA1B_MOUSE
|
||||||
| NC score | 0.019327 (rank : 36) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01901 | Gene names | H2-K1, H2-K | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class I histocompatibility antigen, K-B alpha chain precursor (H- 2K(B)). | |||||
|
HA1D_MOUSE
|
||||||
| NC score | 0.017170 (rank : 37) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01902 | Gene names | H2-K1, H2-K | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class I histocompatibility antigen, K-D alpha chain precursor (H- 2K(D)). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.015956 (rank : 38) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
HA1L_MOUSE
|
||||||
| NC score | 0.011636 (rank : 39) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01897, Q31195, Q31196, Q31197 | Gene names | H2-L | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class I histocompatibility antigen, L-D alpha chain precursor. | |||||