Please be patient as the page loads
|
SYCC_MOUSE
|
||||||
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SYCC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.978650 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49589, Q53XI8, Q9HD24, Q9HD25 | Gene names | CARS | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
SYCC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
SYCM_HUMAN
|
||||||
| θ value | 1.15359e-63 (rank : 3) | NC score | 0.850168 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HA77, Q8NI84, Q96IV4 | Gene names | CARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cysteinyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
SYCM_MOUSE
|
||||||
| θ value | 2.84138e-62 (rank : 4) | NC score | 0.845282 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYM8, Q9D6U9 | Gene names | Cars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cysteinyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
EF1B_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 5) | NC score | 0.190518 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24534, Q6IBH9 | Gene names | EEF1B2, EEF1B, EF1B | |||
|
Domain Architecture |
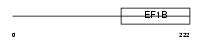
|
|||||
| Description | Elongation factor 1-beta (EF-1-beta). | |||||
|
EF1B_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 6) | NC score | 0.190133 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70251, Q3THP5, Q5SUH1, Q8CHS1, Q99L22, Q9CZD4 | Gene names | Eef1b, Eef1b2 | |||
|
Domain Architecture |
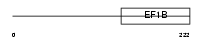
|
|||||
| Description | Elongation factor 1-beta (EF-1-beta). | |||||
|
SYLC_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.150168 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2J5, Q9NSE1 | Gene names | LARS, KIAA1352 | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLC_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.149113 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BMJ2, Q8BKW9 | Gene names | Lars | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.063274 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.052955 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.032731 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.032246 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.032852 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.053800 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
LRC27_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.032876 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.018516 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.021666 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
NOC3L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.044463 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.019133 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
NOC3L_MOUSE
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.044135 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
SYEP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.046042 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.010098 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.026828 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BIN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.030911 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
EF1G_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.032344 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |
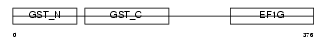
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
SMCA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.016809 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.017547 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.029397 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
CCD19_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.023831 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
DYHC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.022627 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.011379 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
5HT2C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.002253 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28335, Q5VUF8, Q9NP28 | Gene names | HTR2C, HTR1C | |||
|
Domain Architecture |
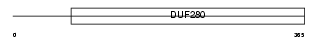
|
|||||
| Description | 5-hydroxytryptamine 2C receptor (5-HT-2C) (Serotonin receptor 2C) (5- HT2C) (5-HTR2C) (5HT-1C). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.021755 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
NUCB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.032313 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
SF04_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.019019 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.017361 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
IF2M_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.015540 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |
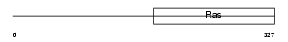
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
IQGA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.016939 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
MCA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.038414 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |
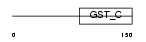
|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.018082 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
RHG08_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.009758 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
SLK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.003642 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.012961 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SPC25_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.021246 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HBM1 | Gene names | SPBC25, SPC25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25 (hSpc25). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.027005 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TPM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.017412 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
CCD38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.017475 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
CJ046_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.026345 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86Y37, Q5XPL7, Q8IY11, Q8N7S4 | Gene names | C10orf46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46. | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.021329 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.024849 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.014346 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
TPM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.017148 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
CILP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.007017 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66K08, Q7TSS0, Q7TSS1, Q8BV01 | Gene names | Cilp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 1 precursor (CILP-1) [Contains: Cartilage intermediate layer protein 1 C1; Cartilage intermediate layer protein 1 C2]. | |||||
|
DJC17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.008221 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
DYHC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.020309 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
EF1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.026065 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |
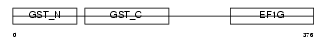
|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
HEAT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.012628 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |
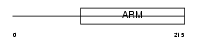
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.004603 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.024833 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.015467 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.015239 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.026765 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.006942 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
EF1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.078717 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29692, Q969J1 | Gene names | EEF1D, EF1D | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-delta (EF-1-delta) (Antigen NY-CO-4). | |||||
|
EF1D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.076354 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57776, Q9CWW2, Q9CWY1, Q9CYD4, Q9CYJ5 | Gene names | Eef1d | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-delta (EF-1-delta). | |||||
|
SYCC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
SYCC_HUMAN
|
||||||
| NC score | 0.978650 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P49589, Q53XI8, Q9HD24, Q9HD25 | Gene names | CARS | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
SYCM_HUMAN
|
||||||
| NC score | 0.850168 (rank : 3) | θ value | 1.15359e-63 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HA77, Q8NI84, Q96IV4 | Gene names | CARS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cysteinyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
SYCM_MOUSE
|
||||||
| NC score | 0.845282 (rank : 4) | θ value | 2.84138e-62 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYM8, Q9D6U9 | Gene names | Cars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cysteinyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
EF1B_HUMAN
|
||||||
| NC score | 0.190518 (rank : 5) | θ value | 1.29631e-06 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24534, Q6IBH9 | Gene names | EEF1B2, EEF1B, EF1B | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-beta (EF-1-beta). | |||||
|
EF1B_MOUSE
|
||||||
| NC score | 0.190133 (rank : 6) | θ value | 1.69304e-06 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70251, Q3THP5, Q5SUH1, Q8CHS1, Q99L22, Q9CZD4 | Gene names | Eef1b, Eef1b2 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-beta (EF-1-beta). | |||||
|
SYLC_HUMAN
|
||||||
| NC score | 0.150168 (rank : 7) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2J5, Q9NSE1 | Gene names | LARS, KIAA1352 | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
SYLC_MOUSE
|
||||||
| NC score | 0.149113 (rank : 8) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BMJ2, Q8BKW9 | Gene names | Lars | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-tRNA synthetase, cytoplasmic (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
EF1D_HUMAN
|
||||||
| NC score | 0.078717 (rank : 9) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29692, Q969J1 | Gene names | EEF1D, EF1D | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-delta (EF-1-delta) (Antigen NY-CO-4). | |||||
|
EF1D_MOUSE
|
||||||
| NC score | 0.076354 (rank : 10) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57776, Q9CWW2, Q9CWY1, Q9CYD4, Q9CYJ5 | Gene names | Eef1d | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-delta (EF-1-delta). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.063274 (rank : 11) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.053800 (rank : 12) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.052955 (rank : 13) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
SYEP_HUMAN
|
||||||
| NC score | 0.046042 (rank : 14) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07814 | Gene names | EPRS, GLNS, PARS, QPRS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
NOC3L_HUMAN
|
||||||
| NC score | 0.044463 (rank : 15) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
NOC3L_MOUSE
|
||||||
| NC score | 0.044135 (rank : 16) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
MCA3_HUMAN
|
||||||
| NC score | 0.038414 (rank : 17) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43324, Q5THS2 | Gene names | EEF1E1 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation elongation factor 1 epsilon-1 (Multisynthetase complex auxiliary component p18) (Elongation factor p18). | |||||
|
LRC27_HUMAN
|
||||||
| NC score | 0.032876 (rank : 18) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.032852 (rank : 19) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.032731 (rank : 20) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
EF1G_MOUSE
|
||||||
| NC score | 0.032344 (rank : 21) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D8N0, Q920C5, Q9CRT5, Q9CSU3, Q9D004 | Gene names | Eef1g | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
NUCB1_MOUSE
|
||||||
| NC score | 0.032313 (rank : 22) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.032246 (rank : 23) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
BIN1_MOUSE
|
||||||
| NC score | 0.030911 (rank : 24) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08539, Q62434 | Gene names | Bin1, Amphl, Sh3p9 | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (SH3-domain-containing protein 9). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.029397 (rank : 25) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.027005 (rank : 26) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.026828 (rank : 27) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.026765 (rank : 28) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
CJ046_HUMAN
|
||||||
| NC score | 0.026345 (rank : 29) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86Y37, Q5XPL7, Q8IY11, Q8N7S4 | Gene names | C10orf46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46. | |||||
|
EF1G_HUMAN
|
||||||
| NC score | 0.026065 (rank : 30) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P26641, Q6PJ62, Q6PK31, Q96CU2, Q9P196 | Gene names | EEF1G, EF1G | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.024849 (rank : 31) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.024833 (rank : 32) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.023831 (rank : 33) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
DYHC_MOUSE
|
||||||
| NC score | 0.022627 (rank : 34) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JHU4 | Gene names | Dync1h1, Dnch1, Dnchc1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.021755 (rank : 35) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.021666 (rank : 36) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.021329 (rank : 37) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
SPC25_HUMAN
|
||||||
| NC score | 0.021246 (rank : 38) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HBM1 | Gene names | SPBC25, SPC25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25 (hSpc25). | |||||
|
DYHC_HUMAN
|
||||||
| NC score | 0.020309 (rank : 39) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 733 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14204, Q6DKQ7, Q8WU28, Q92814, Q9Y4G5 | Gene names | DYNC1H1, DNCH1, DNECL, KIAA0325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) (Dynein heavy chain 1, cytoplasmic 1). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.019133 (rank : 40) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.019019 (rank : 41) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.018516 (rank : 42) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.018082 (rank : 43) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.017547 (rank : 44) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CCD38_HUMAN
|
||||||
| NC score | 0.017475 (rank : 45) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q502W7, Q8N835 | Gene names | CCDC38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 38. | |||||
|
TPM2_HUMAN
|
||||||
| NC score | 0.017412 (rank : 46) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.017361 (rank : 47) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
TPM2_MOUSE
|
||||||
| NC score | 0.017148 (rank : 48) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
IQGA2_HUMAN
|
||||||
| NC score | 0.016939 (rank : 49) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
SMCA1_HUMAN
|
||||||
| NC score | 0.016809 (rank : 50) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
IF2M_MOUSE
|
||||||
| NC score | 0.015540 (rank : 51) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.015467 (rank : 52) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.015239 (rank : 53) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.014346 (rank : 54) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.012961 (rank : 55) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
HEAT3_HUMAN
|
||||||
| NC score | 0.012628 (rank : 56) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.011379 (rank : 57) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.010098 (rank : 58) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
RHG08_HUMAN
|
||||||
| NC score | 0.009758 (rank : 59) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSG0, O75983, O95695, Q96RW1, Q96RW2, Q9HA49, Q9HC46, Q9NVX8, Q9NXL1, Q9UH20 | Gene names | ARHGAP8 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 8. | |||||
|
DJC17_HUMAN
|
||||||
| NC score | 0.008221 (rank : 60) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NVM6 | Gene names | DNAJC17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 17. | |||||
|
CILP1_MOUSE
|
||||||
| NC score | 0.007017 (rank : 61) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q66K08, Q7TSS0, Q7TSS1, Q8BV01 | Gene names | Cilp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 1 precursor (CILP-1) [Contains: Cartilage intermediate layer protein 1 C1; Cartilage intermediate layer protein 1 C2]. | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.006942 (rank : 62) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.004603 (rank : 63) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.003642 (rank : 64) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
5HT2C_HUMAN
|
||||||
| NC score | 0.002253 (rank : 65) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28335, Q5VUF8, Q9NP28 | Gene names | HTR2C, HTR1C | |||
|
Domain Architecture |

|
|||||
| Description | 5-hydroxytryptamine 2C receptor (5-HT-2C) (Serotonin receptor 2C) (5- HT2C) (5-HTR2C) (5HT-1C). | |||||