Please be patient as the page loads
|
SMCA1_MOUSE
|
||||||
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMCA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996003 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SMCA5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.995877 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMCA5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.995972 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 1.47893e-127 (rank : 5) | NC score | 0.941859 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 1.93155e-127 (rank : 6) | NC score | 0.903529 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 4.75757e-126 (rank : 7) | NC score | 0.941440 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 4.02752e-125 (rank : 8) | NC score | 0.937691 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 8.39787e-123 (rank : 9) | NC score | 0.913472 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 1.43246e-122 (rank : 10) | NC score | 0.881164 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 1.58377e-121 (rank : 11) | NC score | 0.878894 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 5.63297e-119 (rank : 12) | NC score | 0.880038 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 5.63297e-119 (rank : 13) | NC score | 0.876171 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 1.02895e-112 (rank : 14) | NC score | 0.934421 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 1.13764e-111 (rank : 15) | NC score | 0.941024 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 3.31004e-111 (rank : 16) | NC score | 0.930269 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 3.31004e-111 (rank : 17) | NC score | 0.925795 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SMRCD_MOUSE
|
||||||
| θ value | 1.49618e-87 (rank : 18) | NC score | 0.950148 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
SMRCD_HUMAN
|
||||||
| θ value | 4.35321e-87 (rank : 19) | NC score | 0.950015 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 1.45253e-74 (rank : 20) | NC score | 0.939042 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
RA54B_HUMAN
|
||||||
| θ value | 2.17053e-70 (rank : 21) | NC score | 0.933636 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
INOC1_HUMAN
|
||||||
| θ value | 4.52582e-68 (rank : 22) | NC score | 0.917896 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
INOC1_MOUSE
|
||||||
| θ value | 7.71989e-68 (rank : 23) | NC score | 0.917968 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
RA54B_MOUSE
|
||||||
| θ value | 1.71981e-67 (rank : 24) | NC score | 0.932763 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | 6.53529e-67 (rank : 25) | NC score | 0.938595 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
RAD54_MOUSE
|
||||||
| θ value | 9.45883e-58 (rank : 26) | NC score | 0.926375 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
RAD54_HUMAN
|
||||||
| θ value | 1.23536e-57 (rank : 27) | NC score | 0.926930 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.02596e-49 (rank : 28) | NC score | 0.822225 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 1.04822e-48 (rank : 29) | NC score | 0.829406 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
TTF2_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 30) | NC score | 0.859726 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 5.0505e-35 (rank : 31) | NC score | 0.818380 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 32) | NC score | 0.838373 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 33) | NC score | 0.844496 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 34) | NC score | 0.695336 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 35) | NC score | 0.676624 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
SMRA3_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 36) | NC score | 0.720769 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
SMRA3_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 37) | NC score | 0.722299 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 38) | NC score | 0.102197 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 39) | NC score | 0.097459 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 40) | NC score | 0.097702 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 41) | NC score | 0.094138 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 42) | NC score | 0.092880 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 43) | NC score | 0.096465 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.086184 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 45) | NC score | 0.061129 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 46) | NC score | 0.061153 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 47) | NC score | 0.011670 (rank : 161) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.061019 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 49) | NC score | 0.012303 (rank : 159) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 50) | NC score | 0.079111 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 51) | NC score | 0.022615 (rank : 136) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 52) | NC score | 0.040106 (rank : 129) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 53) | NC score | 0.016752 (rank : 142) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
LRSM1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 54) | NC score | 0.017900 (rank : 140) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80ZI6 | Gene names | Lrsam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.016783 (rank : 141) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
TNNT2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 56) | NC score | 0.036787 (rank : 130) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 57) | NC score | 0.010711 (rank : 164) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 58) | NC score | 0.003693 (rank : 179) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.053477 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 60) | NC score | 0.053477 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.014870 (rank : 152) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.032244 (rank : 131) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.012394 (rank : 158) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.054807 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.052757 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.052757 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
LBN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.020398 (rank : 137) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K1G2, Q8BRF3 | Gene names | Evc2, Lbn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.015573 (rank : 150) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
UT14A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.024109 (rank : 133) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.015797 (rank : 147) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.059094 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.074883 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.016338 (rank : 143) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 74) | NC score | 0.010263 (rank : 166) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.813845 (rank : 75) | NC score | 0.016250 (rank : 144) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.022700 (rank : 135) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.007325 (rank : 172) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
TNNT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.025512 (rank : 132) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.055051 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
FOXD4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.002943 (rank : 180) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60688, Q61573 | Gene names | Foxd4, Fkh2, Fkhl9, Freac5 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D4 (Forkhead-related protein FKHL9) (Forkhead- related transcription factor 5) (FREAC-5) (Transcription factor FKH- 2). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.071258 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.055333 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.016247 (rank : 145) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
WEE1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | -0.000938 (rank : 182) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47810, Q9EPL7 | Gene names | Wee1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.010169 (rank : 167) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.044839 (rank : 127) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.019971 (rank : 138) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
SH3K1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.008204 (rank : 170) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
ZN628_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | -0.004637 (rank : 184) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
GRK4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | -0.001447 (rank : 183) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P32298, O00641, O00642, Q13293, Q13294, Q13295, Q14453, Q14725, Q15313, Q15314, Q15315, Q15316 | Gene names | GRK4, GPRK2L, GPRK4 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-coupled receptor kinase 4 (EC 2.7.11.16) (G protein-coupled receptor kinase GRK4) (ITI1). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.058210 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.002093 (rank : 181) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
UB7I1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.010700 (rank : 165) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
ZN294_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.011666 (rank : 162) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.009863 (rank : 168) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.005247 (rank : 176) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.015780 (rank : 148) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SH21B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.009122 (rank : 169) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35324, Q8C268 | Gene names | Sh2d1b, Eat2, Sh2d1b1 | |||
|
Domain Architecture |
|
|||||
| Description | SH2 domain protein 1B (EWS/FLI1-activated transcript 2) (EAT-2). | |||||
|
SYCC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.012961 (rank : 155) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
UBF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.015718 (rank : 149) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
ACAD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.004357 (rank : 178) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.041831 (rank : 128) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.011660 (rank : 163) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.006328 (rank : 173) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.005511 (rank : 174) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.013855 (rank : 153) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.015933 (rank : 146) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
UT14A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.011699 (rank : 160) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
VATE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.008078 (rank : 171) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50518 | Gene names | Atp6v1e1, Atp6e | |||
|
Domain Architecture |
|
|||||
| Description | Vacuolar ATP synthase subunit E (EC 3.6.3.14) (V-ATPase E subunit) (Vacuolar proton pump E subunit) (V-ATPase 31 kDa subunit) (P31). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.005385 (rank : 175) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.004477 (rank : 177) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.013716 (rank : 154) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.015564 (rank : 151) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.012488 (rank : 157) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.012625 (rank : 156) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.022946 (rank : 134) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
TNNT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.018222 (rank : 139) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P45378, Q12975, Q12976, Q12977, Q12978, Q86TH6 | Gene names | TNNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT) (Beta TnTF). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.099356 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.101117 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.073568 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.073065 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.078926 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053153 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.052845 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.059732 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.092603 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050199 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.067941 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.067941 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.059902 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.062993 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.082320 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.073497 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.059668 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.062993 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
DD19A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.057366 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.057341 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.057236 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.050095 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.053908 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.055729 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.061187 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050357 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.066476 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.073841 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.064303 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.065803 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LY10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.067309 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051972 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.051569 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.066944 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.060307 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.062217 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.064220 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.055403 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.085166 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.086909 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.087952 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.089086 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.052377 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.051453 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.093267 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.093485 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.051236 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.053504 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.089127 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.054764 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054901 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.077541 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SP110_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.076030 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.057444 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.071015 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.067175 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.070607 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.062001 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.069532 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.071784 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.071992 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.060215 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.050916 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.054462 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.050256 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.082142 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.079602 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SMCA1_HUMAN
|
||||||
| NC score | 0.996003 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA5_MOUSE
|
||||||
| NC score | 0.995972 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMCA5_HUMAN
|
||||||
| NC score | 0.995877 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMRCD_MOUSE
|
||||||
| NC score | 0.950148 (rank : 5) | θ value | 1.49618e-87 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
SMRCD_HUMAN
|
||||||
| NC score | 0.950015 (rank : 6) | θ value | 4.35321e-87 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.941859 (rank : 7) | θ value | 1.47893e-127 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.941440 (rank : 8) | θ value | 4.75757e-126 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.941024 (rank : 9) | θ value | 1.13764e-111 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.939042 (rank : 10) | θ value | 1.45253e-74 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.938595 (rank : 11) | θ value | 6.53529e-67 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.937691 (rank : 12) | θ value | 4.02752e-125 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.934421 (rank : 13) | θ value | 1.02895e-112 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
RA54B_HUMAN
|
||||||
| NC score | 0.933636 (rank : 14) | θ value | 2.17053e-70 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
RA54B_MOUSE
|
||||||
| NC score | 0.932763 (rank : 15) | θ value | 1.71981e-67 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.930269 (rank : 16) | θ value | 3.31004e-111 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
RAD54_HUMAN
|
||||||
| NC score | 0.926930 (rank : 17) | θ value | 1.23536e-57 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
RAD54_MOUSE
|
||||||
| NC score | 0.926375 (rank : 18) | θ value | 9.45883e-58 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.925795 (rank : 19) | θ value | 3.31004e-111 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
INOC1_MOUSE
|
||||||
| NC score | 0.917968 (rank : 20) | θ value | 7.71989e-68 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
INOC1_HUMAN
|
||||||
| NC score | 0.917896 (rank : 21) | θ value | 4.52582e-68 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.913472 (rank : 22) | θ value | 8.39787e-123 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.903529 (rank : 23) | θ value | 1.93155e-127 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.881164 (rank : 24) | θ value | 1.43246e-122 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.880038 (rank : 25) | θ value | 5.63297e-119 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.878894 (rank : 26) | θ value | 1.58377e-121 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.876171 (rank : 27) | θ value | 5.63297e-119 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.859726 (rank : 28) | θ value | 2.34062e-40 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.844496 (rank : 29) | θ value | 5.06226e-27 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.838373 (rank : 30) | θ value | 1.57365e-28 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.829406 (rank : 31) | θ value | 1.04822e-48 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.822225 (rank : 32) | θ value | 8.02596e-49 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.818380 (rank : 33) | θ value | 5.0505e-35 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
SMRA3_MOUSE
|
||||||
| NC score | 0.722299 (rank : 34) | θ value | 1.74391e-19 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
SMRA3_HUMAN
|
||||||
| NC score | 0.720769 (rank : 35) | θ value | 3.51386e-20 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.695336 (rank : 36) | θ value | 1.12775e-26 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.676624 (rank : 37) | θ value | 3.28125e-26 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.102197 (rank : 38) | θ value | 0.000786445 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.101117 (rank : 39) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.099356 (rank : 40) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.097702 (rank : 41) | θ value | 0.00175202 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.097459 (rank : 42) | θ value | 0.00175202 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.096465 (rank : 43) | θ value | 0.00869519 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.094138 (rank : 44) | θ value | 0.00390308 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.093485 (rank : 45) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.093267 (rank : 46) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.092880 (rank : 47) | θ value | 0.00869519 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.092603 (rank : 48) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.089127 (rank : 49) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.089086 (rank : 50) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.087952 (rank : 51) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.086909 (rank : 52) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.086184 (rank : 53) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.085166 (rank : 54) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.082320 (rank : 55) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.082142 (rank : 56) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.079602 (rank : 57) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.079111 (rank : 58) | θ value | 0.0961366 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.078926 (rank : 59) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.077541 (rank : 60) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.076030 (rank : 61) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.074883 (rank : 62) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.073841 (rank : 63) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.073568 (rank : 64) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.073497 (rank : 65) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.073065 (rank : 66) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.071992 (rank : 67) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.071784 (rank : 68) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.071258 (rank : 69) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.071015 (rank : 70) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.070607 (rank : 71) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.069532 (rank : 72) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.067941 (rank : 73) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.067941 (rank : 74) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.067309 (rank : 75) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.067175 (rank : 76) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.066944 (rank : 77) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.066476 (rank : 78) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.065803 (rank : 79) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.064303 (rank : 80) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.064220 (rank : 81) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.062993 (rank : 82) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.062993 (rank : 83) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.062217 (rank : 84) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.062001 (rank : 85) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.061187 (rank : 86) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.061153 (rank : 87) | θ value | 0.0193708 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.061129 (rank : 88) | θ value | 0.0193708 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.061019 (rank : 89) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.060307 (rank : 90) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.060215 (rank : 91) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.059902 (rank : 92) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.059732 (rank : 93) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.059668 (rank : 94) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.059094 (rank : 95) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.058210 (rank : 96) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.057444 (rank : 97) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.057366 (rank : 98) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.057341 (rank : 99) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.057236 (rank : 100) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.055729 (rank : 101) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.055403 (rank : 102) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.055333 (rank : 103) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.055051 (rank : 104) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.054901 (rank : 105) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.054807 (rank : 106) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.054764 (rank : 107) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.054462 (rank : 108) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.053908 (rank : 109) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.053504 (rank : 110) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.053477 (rank : 111) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.053477 (rank : 112) | θ value | 0.365318 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.053153 (rank : 113) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.052845 (rank : 114) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.052757 (rank : 115) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.052757 (rank : 116) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.052377 (rank : 117) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.051972 (rank : 118) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.051569 (rank : 119) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.051453 (rank : 120) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.051236 (rank : 121) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.050916 (rank : 122) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.050357 (rank : 123) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.050256 (rank : 124) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.050199 (rank : 125) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.050095 (rank : 126) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.044839 (rank : 127) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.041831 (rank : 128) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.040106 (rank : 129) | θ value | 0.0961366 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TNNT2_HUMAN
|
||||||
| NC score | 0.036787 (rank : 130) | θ value | 0.163984 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.032244 (rank : 131) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TNNT2_MOUSE
|
||||||
| NC score | 0.025512 (rank : 132) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P50752, Q64360, Q64377 | Gene names | Tnnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
UT14A_MOUSE
|
||||||
| NC score | 0.024109 (rank : 133) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.022946 (rank : 134) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.022700 (rank : 135) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.022615 (rank : 136) | θ value | 0.0961366 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
LBN_MOUSE
|
||||||
| NC score | 0.020398 (rank : 137) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K1G2, Q8BRF3 | Gene names | Evc2, Lbn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.019971 (rank : 138) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
TNNT3_HUMAN
|
||||||
| NC score | 0.018222 (rank : 139) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P45378, Q12975, Q12976, Q12977, Q12978, Q86TH6 | Gene names | TNNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT) (Beta TnTF). | |||||
|
LRSM1_MOUSE
|
||||||
| NC score | 0.017900 (rank : 140) | θ value | 0.163984 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80ZI6 | Gene names | Lrsam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.016783 (rank : 141) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.016752 (rank : 142) | θ value | 0.125558 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.016338 (rank : 143) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.016250 (rank : 144) | θ value | 0.813845 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.016247 (rank : 145) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.015933 (rank : 146) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.015797 (rank : 147) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.015780 (rank : 148) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
UBF1_MOUSE
|
||||||
| NC score | 0.015718 (rank : 149) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P25976 | Gene names | Ubtf, Tcfubf, Ubf-1, Ubf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.015573 (rank : 150) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.015564 (rank : 151) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.014870 (rank : 152) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.013855 (rank : 153) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.013716 (rank : 154) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SYCC_MOUSE
|
||||||
| NC score | 0.012961 (rank : 155) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ER72, O88303, Q8BP81, Q8CAY7, Q8CCE3, Q8K0S4, Q9ER68 | Gene names | Cars | |||
|
Domain Architecture |

|
|||||
| Description | Cysteinyl-tRNA synthetase, cytoplasmic (EC 6.1.1.16) (Cysteine--tRNA ligase) (CysRS). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.012625 (rank : 156) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.012488 (rank : 157) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.012394 (rank : 158) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.012303 (rank : 159) | θ value | 0.0736092 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
UT14A_HUMAN
|
||||||
| NC score | 0.011699 (rank : 160) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.011670 (rank : 161) | θ value | 0.0330416 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ZN294_HUMAN
|
||||||
| NC score | 0.011666 (rank : 162) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.011660 (rank : 163) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.010711 (rank : 164) | θ value | 0.279714 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
UB7I1_HUMAN
|
||||||
| NC score | 0.010700 (rank : 165) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NWF9, Q6Y691, Q75ML7, Q7Z2H7, Q7Z7C1, Q8NHW7, Q9NYT1 | Gene names | UBCE7IP1, ZIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin ligase TRIAD3 (EC 6.3.2.-) (Ubiquitin-conjugating enzyme 7-interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) (Triad domain-containing protein 3). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.010263 (rank : 166) | θ value | 0.813845 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.010169 (rank : 167) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.009863 (rank : 168) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
SH21B_MOUSE
|
||||||
| NC score | 0.009122 (rank : 169) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35324, Q8C268 | Gene names | Sh2d1b, Eat2, Sh2d1b1 | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 1B (EWS/FLI1-activated transcript 2) (EAT-2). | |||||
|
SH3K1_MOUSE
|
||||||
| NC score | 0.008204 (rank : 170) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
VATE_MOUSE
|
||||||
| NC score | 0.008078 (rank : 171) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50518 | Gene names | Atp6v1e1, Atp6e | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase subunit E (EC 3.6.3.14) (V-ATPase E subunit) (Vacuolar proton pump E subunit) (V-ATPase 31 kDa subunit) (P31). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.007325 (rank : 172) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.006328 (rank : 173) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.005511 (rank : 174) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.005385 (rank : 175) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.005247 (rank : 176) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.004477 (rank : 177) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
ACAD9_HUMAN
|
||||||
| NC score | 0.004357 (rank : 178) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H845, Q8WXX3 | Gene names | ACAD9 | |||
|
Domain Architecture |
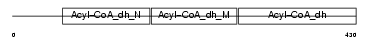
|
|||||
| Description | Acyl-CoA dehydrogenase family member 9, mitochondrial precursor (EC 1.3.99.-) (ACAD-9). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.003693 (rank : 179) | θ value | 0.279714 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
FOXD4_MOUSE
|
||||||
| NC score | 0.002943 (rank : 180) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60688, Q61573 | Gene names | Foxd4, Fkh2, Fkhl9, Freac5 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D4 (Forkhead-related protein FKHL9) (Forkhead- related transcription factor 5) (FREAC-5) (Transcription factor FKH- 2). | |||||
|
SN_MOUSE
|
||||||
| NC score | 0.002093 (rank : 181) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
WEE1_MOUSE
|
||||||
| NC score | -0.000938 (rank : 182) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47810, Q9EPL7 | Gene names | Wee1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase). | |||||
|
GRK4_HUMAN
|
||||||
| NC score | -0.001447 (rank : 183) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P32298, O00641, O00642, Q13293, Q13294, Q13295, Q14453, Q14725, Q15313, Q15314, Q15315, Q15316 | Gene names | GRK4, GPRK2L, GPRK4 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-coupled receptor kinase 4 (EC 2.7.11.16) (G protein-coupled receptor kinase GRK4) (ITI1). | |||||
|
ZN628_HUMAN
|
||||||
| NC score | -0.004637 (rank : 184) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||