Please be patient as the page loads
|
GRP75_HUMAN
|
||||||
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRP75_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998084 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP78_MOUSE
|
||||||
| θ value | 3.98107e-165 (rank : 3) | NC score | 0.975791 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
GRP78_HUMAN
|
||||||
| θ value | 8.86892e-165 (rank : 4) | NC score | 0.975880 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
HSP71_HUMAN
|
||||||
| θ value | 3.99958e-149 (rank : 5) | NC score | 0.982377 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HS70L_HUMAN
|
||||||
| θ value | 7.54289e-148 (rank : 6) | NC score | 0.982934 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HSP7C_MOUSE
|
||||||
| θ value | 1.68038e-147 (rank : 7) | NC score | 0.978184 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HS70L_MOUSE
|
||||||
| θ value | 2.19465e-147 (rank : 8) | NC score | 0.983547 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HSP7C_HUMAN
|
||||||
| θ value | 2.19465e-147 (rank : 9) | NC score | 0.978750 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP76_HUMAN
|
||||||
| θ value | 1.20424e-145 (rank : 10) | NC score | 0.981934 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP72_HUMAN
|
||||||
| θ value | 2.68277e-145 (rank : 11) | NC score | 0.981520 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP72_MOUSE
|
||||||
| θ value | 3.50381e-145 (rank : 12) | NC score | 0.981658 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HS70A_MOUSE
|
||||||
| θ value | 1.92261e-143 (rank : 13) | NC score | 0.981830 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70B_MOUSE
|
||||||
| θ value | 1.92261e-143 (rank : 14) | NC score | 0.981857 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
STCH_MOUSE
|
||||||
| θ value | 2.93355e-67 (rank : 15) | NC score | 0.964564 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_HUMAN
|
||||||
| θ value | 2.32439e-64 (rank : 16) | NC score | 0.963694 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_MOUSE
|
||||||
| θ value | 9.76566e-63 (rank : 17) | NC score | 0.906966 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 1.27544e-62 (rank : 18) | NC score | 0.899580 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HS74L_MOUSE
|
||||||
| θ value | 4.53632e-60 (rank : 19) | NC score | 0.903076 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 7.73779e-60 (rank : 20) | NC score | 0.901015 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 1.51013e-55 (rank : 21) | NC score | 0.889295 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
HSP74_HUMAN
|
||||||
| θ value | 4.39379e-55 (rank : 22) | NC score | 0.901848 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 23) | NC score | 0.871118 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.026845 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.038124 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
HS12B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.159136 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.037962 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
BAP31_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.050502 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51572, Q13836, Q96CF0 | Gene names | BCAP31, BAP31, DXS1357E | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 31 (BCR-associated protein Bap31) (p28 Bap31) (Protein CDM) (6C6-AG tumor-associated antigen). | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.038858 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
HS12A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 30) | NC score | 0.063769 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43301 | Gene names | HSPA12A, KIAA0417 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12A. | |||||
|
HS12A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.063851 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K0U4, Q3UQZ8 | Gene names | Hspa12a | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12A. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.029367 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
BAP31_MOUSE
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.052158 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61335, Q9D0E9, Q9D8G7 | Gene names | Bcap31, Bap31 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 31 (BCR-associated protein Bap31) (p28 Bap31). | |||||
|
HS12B_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.107881 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.055626 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.035785 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.021905 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.044044 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.030963 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.046109 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.009751 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.025909 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.033001 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.014766 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.042292 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.026930 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
HOME1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.044179 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |
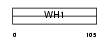
|
|||||
| Description | Homer protein homolog 1. | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.010106 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.041458 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.011778 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
STIM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.019977 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P246, Q96BF1, Q9BQH2, Q9H8R1 | Gene names | STIM2, KIAA1482 | |||
|
Domain Architecture |
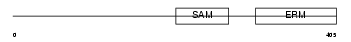
|
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
2AAB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.016325 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30154, O75620 | Gene names | PPP2R1B | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.008343 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
FMOD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.005581 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06828, Q15331 | Gene names | FMOD, FM | |||
|
Domain Architecture |

|
|||||
| Description | Fibromodulin precursor (FM) (Collagen-binding 59 kDa protein) (Keratan sulfate proteoglycan fibromodulin) (KSPG fibromodulin). | |||||
|
FMOD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.005764 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50608 | Gene names | Fmod | |||
|
Domain Architecture |

|
|||||
| Description | Fibromodulin precursor (FM) (Collagen-binding 59 kDa protein) (Keratan sulfate proteoglycan fibromodulin) (KSPG fibromodulin). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.021828 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.027877 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.026127 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.020707 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.040752 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.023392 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
STIM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.019259 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P83093, Q69ZI3, Q80VF4 | Gene names | Stim2, Kiaa1482 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
ABCF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.008879 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
APOL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.013878 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95236, Q9BQ82, Q9BQA3 | Gene names | APOL3 | |||
|
Domain Architecture |
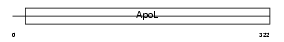
|
|||||
| Description | Apolipoprotein-L3 (Apolipoprotein L-III) (ApoL-III) (TNF-inducible protein CG12-1) (CG12_1). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.011148 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.030896 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.024292 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.036025 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.034425 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.023385 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.020427 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.010463 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.069021 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.019774 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CFDP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.016675 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
MED12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.014505 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.036760 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.035979 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.026602 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.025799 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.023769 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
REST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.029874 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.008955 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.012271 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
EFCB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.006145 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
GCP60_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.018005 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.033919 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.010194 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.012957 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.020828 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.020103 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.008009 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.003291 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.011272 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.009602 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
RAB3I_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.017018 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.004477 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
ANR45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.215856 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.163260 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
COVA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.053807 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
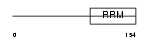
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.059548 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.998084 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
HS70L_MOUSE
|
||||||
| NC score | 0.983547 (rank : 3) | θ value | 2.19465e-147 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HS70L_HUMAN
|
||||||
| NC score | 0.982934 (rank : 4) | θ value | 7.54289e-148 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HSP71_HUMAN
|
||||||
| NC score | 0.982377 (rank : 5) | θ value | 3.99958e-149 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HSP76_HUMAN
|
||||||
| NC score | 0.981934 (rank : 6) | θ value | 1.20424e-145 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HS70B_MOUSE
|
||||||
| NC score | 0.981857 (rank : 7) | θ value | 1.92261e-143 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70A_MOUSE
|
||||||
| NC score | 0.981830 (rank : 8) | θ value | 1.92261e-143 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HSP72_MOUSE
|
||||||
| NC score | 0.981658 (rank : 9) | θ value | 3.50381e-145 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP72_HUMAN
|
||||||
| NC score | 0.981520 (rank : 10) | θ value | 2.68277e-145 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP7C_HUMAN
|
||||||
| NC score | 0.978750 (rank : 11) | θ value | 2.19465e-147 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| NC score | 0.978184 (rank : 12) | θ value | 1.68038e-147 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.975880 (rank : 13) | θ value | 8.86892e-165 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| NC score | 0.975791 (rank : 14) | θ value | 3.98107e-165 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
STCH_MOUSE
|
||||||
| NC score | 0.964564 (rank : 15) | θ value | 2.93355e-67 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_HUMAN
|
||||||
| NC score | 0.963694 (rank : 16) | θ value | 2.32439e-64 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_MOUSE
|
||||||
| NC score | 0.906966 (rank : 17) | θ value | 9.76566e-63 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
HS74L_MOUSE
|
||||||
| NC score | 0.903076 (rank : 18) | θ value | 4.53632e-60 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_HUMAN
|
||||||
| NC score | 0.901848 (rank : 19) | θ value | 4.39379e-55 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.901015 (rank : 20) | θ value | 7.73779e-60 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.899580 (rank : 21) | θ value | 1.27544e-62 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.889295 (rank : 22) | θ value | 1.51013e-55 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.871118 (rank : 23) | θ value | 2.86122e-38 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| NC score | 0.215856 (rank : 24) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR45_MOUSE
|
||||||
| NC score | 0.163260 (rank : 25) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| NC score | 0.159136 (rank : 26) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
HS12B_HUMAN
|
||||||
| NC score | 0.107881 (rank : 27) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.069021 (rank : 28) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
HS12A_MOUSE
|
||||||
| NC score | 0.063851 (rank : 29) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K0U4, Q3UQZ8 | Gene names | Hspa12a | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12A. | |||||
|
HS12A_HUMAN
|
||||||
| NC score | 0.063769 (rank : 30) | θ value | 0.0736092 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43301 | Gene names | HSPA12A, KIAA0417 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12A. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.059548 (rank : 31) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.055626 (rank : 32) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.053807 (rank : 33) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
BAP31_MOUSE
|
||||||
| NC score | 0.052158 (rank : 34) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61335, Q9D0E9, Q9D8G7 | Gene names | Bcap31, Bap31 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 31 (BCR-associated protein Bap31) (p28 Bap31). | |||||
|
BAP31_HUMAN
|
||||||
| NC score | 0.050502 (rank : 35) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51572, Q13836, Q96CF0 | Gene names | BCAP31, BAP31, DXS1357E | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 31 (BCR-associated protein Bap31) (p28 Bap31) (Protein CDM) (6C6-AG tumor-associated antigen). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.046109 (rank : 36) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
HOME1_HUMAN
|
||||||
| NC score | 0.044179 (rank : 37) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1. | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.044044 (rank : 38) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.042292 (rank : 39) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.041458 (rank : 40) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.040752 (rank : 41) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.038858 (rank : 42) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.038124 (rank : 43) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.037962 (rank : 44) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.036760 (rank : 45) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.036025 (rank : 46) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.035979 (rank : 47) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.035785 (rank : 48) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.034425 (rank : 49) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.033919 (rank : 50) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.033001 (rank : 51) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.030963 (rank : 52) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.030896 (rank : 53) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.029874 (rank : 54) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.029367 (rank : 55) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.027877 (rank : 56) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.026930 (rank : 57) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.026845 (rank : 58) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.026602 (rank : 59) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.026127 (rank : 60) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.025909 (rank : 61) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.025799 (rank : 62) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.024292 (rank : 63) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.023769 (rank : 64) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.023392 (rank : 65) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.023385 (rank : 66) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.021905 (rank : 67) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.021828 (rank : 68) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.020828 (rank : 69) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.020707 (rank : 70) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.020427 (rank : 71) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.020103 (rank : 72) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
STIM2_HUMAN
|
||||||
| NC score | 0.019977 (rank : 73) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P246, Q96BF1, Q9BQH2, Q9H8R1 | Gene names | STIM2, KIAA1482 | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.019774 (rank : 74) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
STIM2_MOUSE
|
||||||
| NC score | 0.019259 (rank : 75) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P83093, Q69ZI3, Q80VF4 | Gene names | Stim2, Kiaa1482 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
GCP60_MOUSE
|
||||||
| NC score | 0.018005 (rank : 76) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
RAB3I_HUMAN
|
||||||
| NC score | 0.017018 (rank : 77) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
CFDP1_HUMAN
|
||||||
| NC score | 0.016675 (rank : 78) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UEE9, O00393, O00404, Q9UEF0, Q9UEF1, Q9UEF8 | Gene names | CFDP1, BCNT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Craniofacial development protein 1 (Bucentaur). | |||||
|
2AAB_HUMAN
|
||||||
| NC score | 0.016325 (rank : 79) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30154, O75620 | Gene names | PPP2R1B | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.014766 (rank : 80) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
MED12_HUMAN
|
||||||
| NC score | 0.014505 (rank : 81) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q93074, O15410, O75557, Q9UHV6, Q9UND7 | Gene names | MED12, ARC240, CAGH45, HOPA, KIAA0192, TNRC11, TRAP230 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 12 (Thyroid hormone receptor-associated protein complex 230 kDa component) (Trap230) (Activator-recruited cofactor 240 kDa component) (ARC240) (CAG repeat protein 45) (OPA-containing protein) (Trinucleotide repeat-containing gene 11 protein). | |||||
|
APOL3_HUMAN
|
||||||
| NC score | 0.013878 (rank : 82) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95236, Q9BQ82, Q9BQA3 | Gene names | APOL3 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein-L3 (Apolipoprotein L-III) (ApoL-III) (TNF-inducible protein CG12-1) (CG12_1). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.012957 (rank : 83) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.012271 (rank : 84) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.011778 (rank : 85) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.011272 (rank : 86) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.011148 (rank : 87) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.010463 (rank : 88) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.010194 (rank : 89) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.010106 (rank : 90) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.009751 (rank : 91) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.009602 (rank : 92) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.008955 (rank : 93) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
ABCF1_MOUSE
|
||||||
| NC score | 0.008879 (rank : 94) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P542, Q6NV71 | Gene names | Abcf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1. | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.008343 (rank : 95) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.008009 (rank : 96) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
EFCB1_MOUSE
|
||||||
| NC score | 0.006145 (rank : 97) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
FMOD_MOUSE
|
||||||
| NC score | 0.005764 (rank : 98) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50608 | Gene names | Fmod | |||
|
Domain Architecture |

|
|||||
| Description | Fibromodulin precursor (FM) (Collagen-binding 59 kDa protein) (Keratan sulfate proteoglycan fibromodulin) (KSPG fibromodulin). | |||||
|
FMOD_HUMAN
|
||||||
| NC score | 0.005581 (rank : 99) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06828, Q15331 | Gene names | FMOD, FM | |||
|
Domain Architecture |

|
|||||
| Description | Fibromodulin precursor (FM) (Collagen-binding 59 kDa protein) (Keratan sulfate proteoglycan fibromodulin) (KSPG fibromodulin). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.004477 (rank : 100) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.003291 (rank : 101) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||