Please be patient as the page loads
|
HSP76_HUMAN
|
||||||
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRP78_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.982771 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982759 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
HS70A_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994840 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.994867 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70L_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994075 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HS70L_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.994269 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HSP71_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.995220 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HSP72_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.993665 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP72_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.993691 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP76_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP7C_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.992562 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.991899 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP75_MOUSE
|
||||||
| θ value | 3.16907e-146 (rank : 13) | NC score | 0.982411 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP75_HUMAN
|
||||||
| θ value | 1.20424e-145 (rank : 14) | NC score | 0.981934 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 2.82823e-78 (rank : 15) | NC score | 0.893511 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
HSP74_HUMAN
|
||||||
| θ value | 3.12697e-77 (rank : 16) | NC score | 0.903790 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
STCH_HUMAN
|
||||||
| θ value | 6.52014e-75 (rank : 17) | NC score | 0.963609 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| θ value | 3.23591e-74 (rank : 18) | NC score | 0.963139 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS74L_MOUSE
|
||||||
| θ value | 7.20884e-74 (rank : 19) | NC score | 0.904364 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 1.35953e-72 (rank : 20) | NC score | 0.903248 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 1.96316e-71 (rank : 21) | NC score | 0.900225 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HS105_MOUSE
|
||||||
| θ value | 3.34864e-71 (rank : 22) | NC score | 0.906977 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 2.84797e-54 (rank : 23) | NC score | 0.877027 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 24) | NC score | 0.042742 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.058048 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.050723 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.052906 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.052622 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
ANR45_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.240748 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 30) | NC score | 0.137134 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.032254 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.023304 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.039653 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.070422 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.065900 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.025031 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
PWP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.018898 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LL5, Q9D6T6 | Gene names | Pwp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periodic tryptophan protein 1 homolog. | |||||
|
SLK_MOUSE
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.007704 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.036475 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
PWP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.014916 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13610 | Gene names | PWP1 | |||
|
Domain Architecture |
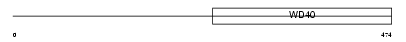
|
|||||
| Description | Periodic tryptophan protein 1 homolog (Keratinocyte protein IEF SSP 9502). | |||||
|
ANR38_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.015332 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P9J5, Q8BV03 | Gene names | Ankrd38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
SLK_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.006307 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
ANR45_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.183071 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
CCD43_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.031482 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96MW1 | Gene names | CCDC43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 43. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.046917 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HS12B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.083208 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.051294 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.027483 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
FLNA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.034082 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
K2C4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.013207 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
HOOK2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.020647 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.052673 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.026182 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
CCD43_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.029475 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CR29, Q6PDT2 | Gene names | Ccdc43, D11Ertd707e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 43. | |||||
|
GCP60_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.024115 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.041139 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.027333 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.039868 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CHCH3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.027654 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CRB9 | Gene names | Chchd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3. | |||||
|
GCP60_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.025330 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.022877 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MYO7A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.028445 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97479 | Gene names | Myo7a, Myo7 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-7A (Myosin VIIa). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.031871 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.029589 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.023422 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.036928 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SAFB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.012353 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.023830 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.032732 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.047277 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.040892 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
TNIP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.025927 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.030109 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.027567 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.004328 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
NEMO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.022635 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
TAF7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.035026 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15545, Q13036 | Gene names | TAF7, TAF2F, TAFII55 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 7 (Transcription initiation factor TFIID 55 kDa subunit) (TAF(II)55) (TAFII-55) (TAFII55). | |||||
|
TAF7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.038573 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1C0 | Gene names | Taf7, Taf2f | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 7 (Transcription initiation factor TFIID 55 kDa subunit) (TAF(II)55) (TAFII-55) (TAFII55). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.027032 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.021443 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.008314 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.035189 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.036987 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.021876 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.033697 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.020100 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
TESK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | -0.002251 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |
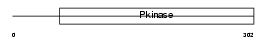
|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.022545 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.028662 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.011519 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.015453 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.024870 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
REST_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.034380 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RFIP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.029113 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.032115 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
TNNT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.033264 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
VPS35_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.011327 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QK1, Q561W2, Q9H016, Q9H096, Q9H4P3, Q9H8J0, Q9NRS7, Q9NVG2, Q9NX80, Q9NZK2 | Gene names | VPS35, MEM3 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (hVPS35) (Maternal-embryonic 3). | |||||
|
VPS35_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.011327 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQH3, Q61123 | Gene names | Vps35, Mem3 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (Maternal-embryonic 3). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.026631 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.015518 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.016962 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.010652 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
COVA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.055976 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
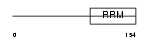
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.028426 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
FLNB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.013560 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
FLNB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.012548 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.046334 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.024812 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.041248 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.042302 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.037014 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.037609 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.040100 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.031033 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.026697 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
RFIP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.022923 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.058652 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SYWM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.035866 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYK1 | Gene names | Wars2 | |||
|
Domain Architecture |
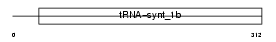
|
|||||
| Description | Tryptophanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) ((Mt)TrpRS). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.030362 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.048543 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.003232 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.001210 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.001602 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.018265 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.024126 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.017607 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
UBXD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.034002 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
UBXD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.031077 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
FA44A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.050848 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
HSP76_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 128 | |
| SwissProt Accessions | P17066, Q8IYK7, Q9BT95 | Gene names | HSPA6, HSP70B' | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 6 (Heat shock 70 kDa protein B'). | |||||
|
HSP71_HUMAN
|
||||||
| NC score | 0.995220 (rank : 2) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P08107, P19790, Q9UQL9, Q9UQM0 | Gene names | HSPA1A, HSPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1 (HSP70.1) (HSP70-1/HSP70-2). | |||||
|
HS70B_MOUSE
|
||||||
| NC score | 0.994867 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P17879, Q61689, Q925V6 | Gene names | Hspa1b, Hcp70.1, Hsp70-1, Hsp70a1, Hspa1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1B (HSP70.1). | |||||
|
HS70A_MOUSE
|
||||||
| NC score | 0.994840 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61696, Q61697, Q7TQD8, Q9QWJ5 | Gene names | Hspa1a, Hsp70-3, Hsp70A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 1A (Heat shock 70 kDa protein 3) (HSP70.3) (Hsp68). | |||||
|
HS70L_MOUSE
|
||||||
| NC score | 0.994269 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16627, O88686, Q61693 | Gene names | Hspa1l, Hsc70t | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa-like protein 1) (Spermatid-specific heat shock protein 70). | |||||
|
HS70L_HUMAN
|
||||||
| NC score | 0.994075 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
HSP72_MOUSE
|
||||||
| NC score | 0.993691 (rank : 7) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
HSP72_HUMAN
|
||||||
| NC score | 0.993665 (rank : 8) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
HSP7C_HUMAN
|
||||||
| NC score | 0.992562 (rank : 9) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P11142, Q9H3R6 | Gene names | HSPA8, HSC70, HSP73, HSPA10 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
HSP7C_MOUSE
|
||||||
| NC score | 0.991899 (rank : 10) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P63017, P08109, P12225, Q62373, Q62374, Q62375 | Gene names | Hspa8, Hsc70, Hsc73 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8). | |||||
|
GRP78_HUMAN
|
||||||
| NC score | 0.982771 (rank : 11) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P11021, Q2EF78, Q9NPF1, Q9UK02 | Gene names | HSPA5, GRP78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+)-binding protein grp78). | |||||
|
GRP78_MOUSE
|
||||||
| NC score | 0.982759 (rank : 12) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P20029, O35642, Q3UFF2, Q61630 | Gene names | Hspa5, Grp78 | |||
|
Domain Architecture |

|
|||||
| Description | 78 kDa glucose-regulated protein precursor (GRP 78) (Heat shock 70 kDa protein 5) (Immunoglobulin heavy chain-binding protein) (BiP). | |||||
|
GRP75_MOUSE
|
||||||
| NC score | 0.982411 (rank : 13) | θ value | 3.16907e-146 (rank : 13) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P38647, Q9CQ05 | Gene names | Hspa9a, Grp75, Hsp74, Hspa9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (P66 MOT) (Mortalin). | |||||
|
GRP75_HUMAN
|
||||||
| NC score | 0.981934 (rank : 14) | θ value | 1.20424e-145 (rank : 14) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P38646, P30036, P31932, Q9BWB7 | Gene names | HSPA9B, GRP75, HSPA9 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) (Peptide-binding protein 74) (PBP74) (Mortalin) (MOT). | |||||
|
STCH_HUMAN
|
||||||
| NC score | 0.963609 (rank : 15) | θ value | 6.52014e-75 (rank : 17) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| NC score | 0.963139 (rank : 16) | θ value | 3.23591e-74 (rank : 18) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
HS105_MOUSE
|
||||||
| NC score | 0.906977 (rank : 17) | θ value | 3.34864e-71 (rank : 22) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61699, Q3TNS2, Q3UIY8, Q62578, Q62579, Q6A0A5, Q8C430, Q8VCW6 | Gene names | Hsph1, Hsp105, Hsp110, Kiaa0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock-related 100 kDa protein E7I) (HSP-E7I) (Heat shock 110 kDa protein) (42 degrees C-HSP). | |||||
|
HS74L_MOUSE
|
||||||
| NC score | 0.904364 (rank : 18) | θ value | 7.20884e-74 (rank : 19) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HSP74_HUMAN
|
||||||
| NC score | 0.903790 (rank : 19) | θ value | 3.12697e-77 (rank : 16) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P34932, O95756 | Gene names | HSPA4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2) (HSP70RY). | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.903248 (rank : 20) | θ value | 1.35953e-72 (rank : 20) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.900225 (rank : 21) | θ value | 1.96316e-71 (rank : 21) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.893511 (rank : 22) | θ value | 2.82823e-78 (rank : 15) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.877027 (rank : 23) | θ value | 2.84797e-54 (rank : 23) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
ANR45_HUMAN
|
||||||
| NC score | 0.240748 (rank : 24) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TZF3, Q6ZST1 | Gene names | ANKRD45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
ANR45_MOUSE
|
||||||
| NC score | 0.183071 (rank : 25) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q810N6 | Gene names | Ankrd45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 45. | |||||
|
HS12B_MOUSE
|
||||||
| NC score | 0.137134 (rank : 26) | θ value | 0.0736092 (rank : 30) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CZJ2 | Gene names | Hspa12b | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
HS12B_HUMAN
|
||||||
| NC score | 0.083208 (rank : 27) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96MM6, Q9BR52 | Gene names | HSPA12B, C20orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock 70 kDa protein 12B. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.070422 (rank : 28) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.065900 (rank : 29) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.058652 (rank : 30) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.058048 (rank : 31) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.055976 (rank : 32) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.052906 (rank : 33) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.052673 (rank : 34) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.052622 (rank : 35) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.051294 (rank : 36) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.050848 (rank : 37) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.050723 (rank : 38) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.048543 (rank : 39) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.047277 (rank : 40) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.046917 (rank : 41) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.046334 (rank : 42) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.042742 (rank : 43) | θ value | 0.0193708 (rank : 24) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.042302 (rank : 44) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.041248 (rank : 45) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.041139 (rank : 46) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.040892 (rank : 47) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.040100 (rank : 48) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.039868 (rank : 49) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.039653 (rank : 50) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
TAF7_MOUSE
|
||||||
| NC score | 0.038573 (rank : 51) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1C0 | Gene names | Taf7, Taf2f | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 7 (Transcription initiation factor TFIID 55 kDa subunit) (TAF(II)55) (TAFII-55) (TAFII55). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.037609 (rank : 52) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.037014 (rank : 53) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.036987 (rank : 54) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.036928 (rank : 55) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.036475 (rank : 56) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
SYWM_MOUSE
|
||||||
| NC score | 0.035866 (rank : 57) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYK1 | Gene names | Wars2 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptophanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.2) (Tryptophan--tRNA ligase) (TrpRS) ((Mt)TrpRS). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.035189 (rank : 58) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TAF7_HUMAN
|
||||||
| NC score | 0.035026 (rank : 59) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15545, Q13036 | Gene names | TAF7, TAF2F, TAFII55 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 7 (Transcription initiation factor TFIID 55 kDa subunit) (TAF(II)55) (TAFII-55) (TAFII55). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.034380 (rank : 60) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
FLNA_HUMAN
|
||||||
| NC score | 0.034082 (rank : 61) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P21333 | Gene names | FLNA, FLN, FLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
UBXD2_HUMAN
|
||||||
| NC score | 0.034002 (rank : 62) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.033697 (rank : 63) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
TNNT2_HUMAN
|
||||||
| NC score | 0.033264 (rank : 64) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P45379, O60214, Q99596, Q99597, Q9UM96 | Gene names | TNNT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, cardiac muscle (TnTc) (Cardiac muscle troponin T) (cTnT). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.032732 (rank : 65) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.032254 (rank : 66) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.032115 (rank : 67) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.031871 (rank : 68) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CCD43_HUMAN
|
||||||
| NC score | 0.031482 (rank : 69) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96MW1 | Gene names | CCDC43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 43. | |||||
|
UBXD2_MOUSE
|
||||||
| NC score | 0.031077 (rank : 70) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.031033 (rank : 71) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.030362 (rank : 72) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.030109 (rank : 73) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.029589 (rank : 74) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
CCD43_MOUSE
|
||||||
| NC score | 0.029475 (rank : 75) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CR29, Q6PDT2 | Gene names | Ccdc43, D11Ertd707e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 43. | |||||
|
RFIP4_HUMAN
|
||||||
| NC score | 0.029113 (rank : 76) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.028662 (rank : 77) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
MYO7A_MOUSE
|
||||||
| NC score | 0.028445 (rank : 78) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97479 | Gene names | Myo7a, Myo7 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-7A (Myosin VIIa). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.028426 (rank : 79) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CHCH3_MOUSE
|
||||||
| NC score | 0.027654 (rank : 80) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9CRB9 | Gene names | Chchd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3. | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.027567 (rank : 81) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.027483 (rank : 82) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.027333 (rank : 83) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.027032 (rank : 84) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.026697 (rank : 85) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.026631 (rank : 86) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.026182 (rank : 87) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TNIP3_HUMAN
|
||||||
| NC score | 0.025927 (rank : 88) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
GCP60_MOUSE
|
||||||
| NC score | 0.025330 (rank : 89) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.025031 (rank : 90) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.024870 (rank : 91) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.024812 (rank : 92) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.024126 (rank : 93) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
GCP60_HUMAN
|
||||||
| NC score | 0.024115 (rank : 94) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.023830 (rank : 95) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.023422 (rank : 96) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.023304 (rank : 97) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
RFIP4_MOUSE
|
||||||
| NC score | 0.022923 (rank : 98) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.022877 (rank : 99) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
NEMO_MOUSE
|
||||||
| NC score | 0.022635 (rank : 100) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O88522 | Gene names | Ikbkg, Nemo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (mFIP-3). | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.022545 (rank : 101) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.021876 (rank : 102) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.021443 (rank : 103) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
HOOK2_MOUSE
|
||||||
| NC score | 0.020647 (rank : 104) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.020100 (rank : 105) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
PWP1_MOUSE
|
||||||
| NC score | 0.018898 (rank : 106) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LL5, Q9D6T6 | Gene names | Pwp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periodic tryptophan protein 1 homolog. | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.018265 (rank : 107) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.017607 (rank : 108) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.016962 (rank : 109) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.015518 (rank : 110) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.015453 (rank : 111) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
ANR38_MOUSE
|
||||||
| NC score | 0.015332 (rank : 112) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 512 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P9J5, Q8BV03 | Gene names | Ankrd38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
PWP1_HUMAN
|
||||||
| NC score | 0.014916 (rank : 113) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13610 | Gene names | PWP1 | |||
|
Domain Architecture |

|
|||||
| Description | Periodic tryptophan protein 1 homolog (Keratinocyte protein IEF SSP 9502). | |||||
|
FLNB_HUMAN
|
||||||
| NC score | 0.013560 (rank : 114) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
K2C4_MOUSE
|
||||||
| NC score | 0.013207 (rank : 115) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
FLNB_MOUSE
|
||||||
| NC score | 0.012548 (rank : 116) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
SAFB2_MOUSE
|
||||||
| NC score | 0.012353 (rank : 117) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.011519 (rank : 118) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
VPS35_MOUSE
|
||||||
| NC score | 0.011327 (rank : 119) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQH3, Q61123 | Gene names | Vps35, Mem3 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (Maternal-embryonic 3). | |||||
|
VPS35_HUMAN
|
||||||
| NC score | 0.011327 (rank : 120) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QK1, Q561W2, Q9H016, Q9H096, Q9H4P3, Q9H8J0, Q9NRS7, Q9NVG2, Q9NX80, Q9NZK2 | Gene names | VPS35, MEM3 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (hVPS35) (Maternal-embryonic 3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.010652 (rank : 121) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.008314 (rank : 122) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.007704 (rank : 123) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.006307 (rank : 124) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.004328 (rank : 125) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.003232 (rank : 126) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
MINK1_MOUSE
|
||||||
| NC score | 0.001602 (rank : 127) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_HUMAN
|
||||||
| NC score | 0.001210 (rank : 128) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
TESK2_MOUSE
|
||||||
| NC score | -0.002251 (rank : 129) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 128 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCT9 | Gene names | Tesk2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity testis-specific protein kinase 2 (EC 2.7.12.1) (Testicular protein kinase 2). | |||||