Please be patient as the page loads
|
SMRCD_HUMAN
|
||||||
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMRCD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
SMRCD_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995177 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | 4.35321e-87 (rank : 3) | NC score | 0.950015 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
SMCA5_HUMAN
|
||||||
| θ value | 7.42547e-87 (rank : 4) | NC score | 0.953079 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMCA5_MOUSE
|
||||||
| θ value | 7.42547e-87 (rank : 5) | NC score | 0.953076 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMCA1_HUMAN
|
||||||
| θ value | 3.11973e-85 (rank : 6) | NC score | 0.951852 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 1.95861e-79 (rank : 7) | NC score | 0.923319 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 1.95861e-79 (rank : 8) | NC score | 0.890076 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 4.36332e-79 (rank : 9) | NC score | 0.927195 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 5.69867e-79 (rank : 10) | NC score | 0.901835 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 1.65806e-78 (rank : 11) | NC score | 0.928637 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 7.20884e-74 (rank : 12) | NC score | 0.927526 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 2.73936e-73 (rank : 13) | NC score | 0.921420 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 6.7473e-72 (rank : 14) | NC score | 0.915801 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 1.50314e-71 (rank : 15) | NC score | 0.859283 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 1.50314e-71 (rank : 16) | NC score | 0.855292 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 1.50314e-71 (rank : 17) | NC score | 0.911070 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 2.83479e-70 (rank : 18) | NC score | 0.859748 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ERCC6_HUMAN
|
||||||
| θ value | 2.32439e-64 (rank : 19) | NC score | 0.938797 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 1.50663e-63 (rank : 20) | NC score | 0.856235 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | 2.17557e-62 (rank : 21) | NC score | 0.944614 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
RA54B_HUMAN
|
||||||
| θ value | 3.47335e-60 (rank : 22) | NC score | 0.936641 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
INOC1_MOUSE
|
||||||
| θ value | 1.45929e-58 (rank : 23) | NC score | 0.920096 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
INOC1_HUMAN
|
||||||
| θ value | 1.90589e-58 (rank : 24) | NC score | 0.919662 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
RA54B_MOUSE
|
||||||
| θ value | 9.78833e-55 (rank : 25) | NC score | 0.932853 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
RAD54_HUMAN
|
||||||
| θ value | 1.61718e-49 (rank : 26) | NC score | 0.929815 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
RAD54_MOUSE
|
||||||
| θ value | 1.36902e-48 (rank : 27) | NC score | 0.928179 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 28) | NC score | 0.829866 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.79215e-40 (rank : 29) | NC score | 0.821313 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
TTF2_HUMAN
|
||||||
| θ value | 3.86705e-35 (rank : 30) | NC score | 0.874938 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
SMRA3_HUMAN
|
||||||
| θ value | 4.27553e-34 (rank : 31) | NC score | 0.764208 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
SMRA3_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 32) | NC score | 0.767395 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 33) | NC score | 0.828509 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 34) | NC score | 0.826536 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 35) | NC score | 0.841009 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 36) | NC score | 0.691484 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 37) | NC score | 0.673361 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 38) | NC score | 0.027549 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 39) | NC score | 0.030887 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
OSTP_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 40) | NC score | 0.040751 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 41) | NC score | 0.027950 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 42) | NC score | 0.040818 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 43) | NC score | 0.010309 (rank : 145) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.038420 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.038513 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.038449 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.017050 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.025603 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
LRC50_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.015749 (rank : 129) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2H9, Q9CVS9 | Gene names | Lrrc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.019098 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.026188 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.023847 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.021590 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.011951 (rank : 141) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.011385 (rank : 144) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.021003 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
ERR3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.004375 (rank : 161) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62508, O75454, O96021, Q68DA0, Q6P274, Q6PK28, Q6TS38, Q9R1F3, Q9UNJ4 | Gene names | ESRRG, ERR3, ERRG2, KIAA0832, NR3B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estrogen-related receptor gamma (Estrogen receptor-related protein 3) (ERR gamma-2). | |||||
|
ERR3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.004375 (rank : 162) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62509, O75454, O96021, Q8CHC9, Q9R1F3 | Gene names | Esrrg, Err3, Kiaa0832, Nr3b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estrogen-related receptor gamma (Estrogen receptor-related protein 3). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.011980 (rank : 139) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.019566 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.015873 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.012639 (rank : 137) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.014996 (rank : 132) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.024396 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
CCD11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.011961 (rank : 140) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CI093_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.011469 (rank : 142) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.008323 (rank : 151) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
LYRIC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.016859 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
ACTN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.004478 (rank : 160) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
AL2SA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.015373 (rank : 130) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | -0.001084 (rank : 167) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.019659 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.016703 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
INADL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.006422 (rank : 156) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.007280 (rank : 153) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
PARP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.009913 (rank : 146) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.013497 (rank : 133) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.016633 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.037496 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
EVC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.012855 (rank : 135) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P57679 | Gene names | EVC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein (DWF-1). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.016725 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
IMMT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.015868 (rank : 127) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CAQ8, Q66JS4, Q7TNE2, Q8C7V1, Q8CCI0, Q8CDA8, Q9D9F6 | Gene names | Immt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin). | |||||
|
LYRIC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.018596 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.012819 (rank : 136) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.016385 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | -0.001818 (rank : 169) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | -0.001501 (rank : 168) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
CM007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.022776 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.049047 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.071374 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.008724 (rank : 150) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.003895 (rank : 164) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.004748 (rank : 159) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
ACTN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.004073 (rank : 163) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.041258 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.023368 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.017820 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CM007_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.028001 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5W0B1, Q8TBY2, Q9H0T2, Q9H8M0 | Gene names | C13orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7. | |||||
|
CT174_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.012865 (rank : 134) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.015138 (rank : 131) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
PDPK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | -0.004600 (rank : 171) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15530 | Gene names | PDPK1, PDK1 | |||
|
Domain Architecture |
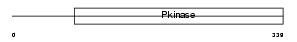
|
|||||
| Description | 3-phosphoinositide-dependent protein kinase 1 (EC 2.7.11.1) (hPDK1). | |||||
|
PDPK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | -0.004591 (rank : 170) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2A0, Q9R1D8, Q9R215 | Gene names | Pdpk1, Pdk1 | |||
|
Domain Architecture |
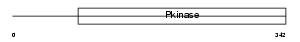
|
|||||
| Description | 3-phosphoinositide-dependent protein kinase 1 (EC 2.7.11.1) (mPDK1). | |||||
|
TPM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.009789 (rank : 147) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.009707 (rank : 148) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.012409 (rank : 138) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.005628 (rank : 158) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
CCD82_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.007266 (rank : 154) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PG04, Q3V462 | Gene names | Ccdc82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
CK035_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.006333 (rank : 157) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q0VET5, Q9CW10 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35 homolog. | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.009369 (rank : 149) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.018868 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.003840 (rank : 166) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.003852 (rank : 165) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
GRAM3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.011387 (rank : 143) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PEM6 | Gene names | Gramd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRAM domain-containing protein 3. | |||||
|
KIF11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.006855 (rank : 155) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P9P6, Q9Z1J0 | Gene names | Kif11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5). | |||||
|
MGRN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.008323 (rank : 152) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.031969 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
UT14A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.015769 (rank : 128) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.092111 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.093929 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.069863 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.066401 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.073376 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.051641 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.056309 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.088646 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.065264 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.065264 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.056612 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.059691 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.078362 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.069805 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.057113 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.060351 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
DDX41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.051108 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.051134 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.050161 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.073478 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.064432 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.066404 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LY10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.063566 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.052026 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.065248 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.057871 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.057943 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.059529 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.053396 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.079252 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.080436 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.081359 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.082217 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.087003 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.086995 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050616 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.083552 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PL10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.053153 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.056365 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.054898 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.072874 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SP110_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.070844 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.054605 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.070477 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.063361 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.066385 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.058479 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.065755 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.067198 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.066957 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.056866 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.052881 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.077487 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.075297 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
SMRCD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9H4L7, Q96SX1, Q9H017, Q9H860, Q9NPU9, Q9ULU7 | Gene names | SMARCAD1, KIAA1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase 1) (hHEL1). | |||||
|
SMRCD_MOUSE
|
||||||
| NC score | 0.995177 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q04692 | Gene names | Smarcad1, Etl1, Kiaa1122 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3.6.1.-) (ATP- dependent helicase SMARCAD1) (Enhancer trap locus homolog 1) (Etl-1). | |||||
|
SMCA5_HUMAN
|
||||||
| NC score | 0.953079 (rank : 3) | θ value | 7.42547e-87 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60264 | Gene names | SMARCA5, SNF2H, WCRF135 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin A5) (Sucrose nonfermenting protein 2 homolog) (hSNF2H). | |||||
|
SMCA5_MOUSE
|
||||||
| NC score | 0.953076 (rank : 4) | θ value | 7.42547e-87 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZW3, Q8C791, Q8CA22, Q8VDG1, Q925M9 | Gene names | Smarca5, Snf2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 5 (EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog) (mSnf2h). | |||||
|
SMCA1_HUMAN
|
||||||
| NC score | 0.951852 (rank : 5) | θ value | 3.11973e-85 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P28370, Q5JV41, Q5JV42 | Gene names | SMARCA1, SNF2L, SNF2L1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.950015 (rank : 6) | θ value | 4.35321e-87 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.944614 (rank : 7) | θ value | 2.17557e-62 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
ERCC6_HUMAN
|
||||||
| NC score | 0.938797 (rank : 8) | θ value | 2.32439e-64 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q03468 | Gene names | ERCC6, CSB | |||
|
Domain Architecture |

|
|||||
| Description | DNA excision repair protein ERCC-6 (EC 3.6.1.-) (ATP-dependent helicase ERCC6) (Cockayne syndrome protein CSB). | |||||
|
RA54B_HUMAN
|
||||||
| NC score | 0.936641 (rank : 9) | θ value | 3.47335e-60 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y620 | Gene names | RAD54B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
RA54B_MOUSE
|
||||||
| NC score | 0.932853 (rank : 10) | θ value | 9.78833e-55 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PFE3 | Gene names | Rad54b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54B (EC 3.6.1.-) (RAD54 homolog B). | |||||
|
RAD54_HUMAN
|
||||||
| NC score | 0.929815 (rank : 11) | θ value | 1.61718e-49 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.928637 (rank : 12) | θ value | 1.65806e-78 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
RAD54_MOUSE
|
||||||
| NC score | 0.928179 (rank : 13) | θ value | 1.36902e-48 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.927526 (rank : 14) | θ value | 7.20884e-74 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.927195 (rank : 15) | θ value | 4.36332e-79 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.923319 (rank : 16) | θ value | 1.95861e-79 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.921420 (rank : 17) | θ value | 2.73936e-73 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
INOC1_MOUSE
|
||||||
| NC score | 0.920096 (rank : 18) | θ value | 1.45929e-58 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZPV2, Q6P7V0, Q6PCP1, Q8C9T7 | Gene names | Inoc1, Kiaa1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-). | |||||
|
INOC1_HUMAN
|
||||||
| NC score | 0.919662 (rank : 19) | θ value | 1.90589e-58 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9ULG1, Q9NTG6 | Gene names | INOC1, KIAA1259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative DNA helicase INO80 complex homolog 1 (EC 3.6.1.-) (hINO80). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.915801 (rank : 20) | θ value | 6.7473e-72 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.911070 (rank : 21) | θ value | 1.50314e-71 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.901835 (rank : 22) | θ value | 5.69867e-79 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.890076 (rank : 23) | θ value | 1.95861e-79 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.874938 (rank : 24) | θ value | 3.86705e-35 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.859748 (rank : 25) | θ value | 2.83479e-70 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.859283 (rank : 26) | θ value | 1.50314e-71 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.856235 (rank : 27) | θ value | 1.50663e-63 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.855292 (rank : 28) | θ value | 1.50314e-71 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.841009 (rank : 29) | θ value | 5.23862e-24 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.829866 (rank : 30) | θ value | 2.50074e-42 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.828509 (rank : 31) | θ value | 6.82597e-32 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.826536 (rank : 32) | θ value | 1.62847e-25 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.821313 (rank : 33) | θ value | 1.79215e-40 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
SMRA3_MOUSE
|
||||||
| NC score | 0.767395 (rank : 34) | θ value | 7.29293e-34 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
SMRA3_HUMAN
|
||||||
| NC score | 0.764208 (rank : 35) | θ value | 4.27553e-34 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.691484 (rank : 36) | θ value | 6.62687e-19 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.673361 (rank : 37) | θ value | 8.65492e-19 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.093929 (rank : 38) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.092111 (rank : 39) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.088646 (rank : 40) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.087003 (rank : 41) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.086995 (rank : 42) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.083552 (rank : 43) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.082217 (rank : 44) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.081359 (rank : 45) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.080436 (rank : 46) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.079252 (rank : 47) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.078362 (rank : 48) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.077487 (rank : 49) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.075297 (rank : 50) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.073478 (rank : 51) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.073376 (rank : 52) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.072874 (rank : 53) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.071374 (rank : 54) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.070844 (rank : 55) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.070477 (rank : 56) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.069863 (rank : 57) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.069805 (rank : 58) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.067198 (rank : 59) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.066957 (rank : 60) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.066404 (rank : 61) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.066401 (rank : 62) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.066385 (rank : 63) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.065755 (rank : 64) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.065264 (rank : 65) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.065264 (rank : 66) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.065248 (rank : 67) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.064432 (rank : 68) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.063566 (rank : 69) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.063361 (rank : 70) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.060351 (rank : 71) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.059691 (rank : 72) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.059529 (rank : 73) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.058479 (rank : 74) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.057943 (rank : 75) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.057871 (rank : 76) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.057113 (rank : 77) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.056866 (rank : 78) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.056612 (rank : 79) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.056365 (rank : 80) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.056309 (rank : 81) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.054898 (rank : 82) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.054605 (rank : 83) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.053396 (rank : 84) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.053153 (rank : 85) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.052881 (rank : 86) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.052026 (rank : 87) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.051641 (rank : 88) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.051134 (rank : 89) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.051108 (rank : 90) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.050616 (rank : 91) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.050161 (rank : 92) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.049047 (rank : 93) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.041258 (rank : 94) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.040818 (rank : 95) | θ value | 0.0563607 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
OSTP_MOUSE
|
||||||
| NC score | 0.040751 (rank : 96) | θ value | 0.0252991 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10923, P19008 | Gene names | Spp1, Eta-1, Op, Spp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Osteopontin precursor (Bone sialoprotein-1) (Secreted phosphoprotein 1) (SPP-1) (Minopontin) (Early T-lymphocyte activation 1 protein) (2AR) (Calcium oxalate crystal growth inhibitor protein). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.038513 (rank : 97) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.038449 (rank : 98) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.038420 (rank : 99) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.037496 (rank : 100) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.031969 (rank : 101) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.030887 (rank : 102) | θ value | 0.0193708 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
CM007_HUMAN
|
||||||
| NC score | 0.028001 (rank : 103) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5W0B1, Q8TBY2, Q9H0T2, Q9H8M0 | Gene names | C13orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7. | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.027950 (rank : 104) | θ value | 0.0431538 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.027549 (rank : 105) | θ value | 0.0193708 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.026188 (rank : 106) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.025603 (rank : 107) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.024396 (rank : 108) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.023847 (rank : 109) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.023368 (rank : 110) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.022776 (rank : 111) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.021590 (rank : 112) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.021003 (rank : 113) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.019659 (rank : 114) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.019566 (rank : 115) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.019098 (rank : 116) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.018868 (rank : 117) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
LYRIC_HUMAN
|
||||||
| NC score | 0.018596 (rank : 118) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.017820 (rank : 119) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.017050 (rank : 120) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
LYRIC_MOUSE
|
||||||
| NC score | 0.016859 (rank : 121) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.016725 (rank : 122) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.016703 (rank : 123) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.016633 (rank : 124) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.016385 (rank : 125) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.015873 (rank : 126) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
IMMT_MOUSE
|
||||||
| NC score | 0.015868 (rank : 127) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CAQ8, Q66JS4, Q7TNE2, Q8C7V1, Q8CCI0, Q8CDA8, Q9D9F6 | Gene names | Immt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial inner membrane protein (Mitofilin). | |||||
|
UT14A_MOUSE
|
||||||
| NC score | 0.015769 (rank : 128) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
LRC50_MOUSE
|
||||||
| NC score | 0.015749 (rank : 129) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2H9, Q9CVS9 | Gene names | Lrrc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
AL2SA_HUMAN
|
||||||
| NC score | 0.015373 (rank : 130) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q53TS8, Q8NCN6, Q96LN4 | Gene names | ALS2CR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 11 protein. | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.015138 (rank : 131) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.014996 (rank : 132) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.013497 (rank : 133) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.012865 (rank : 134) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
EVC_HUMAN
|
||||||
| NC score | 0.012855 (rank : 135) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P57679 | Gene names | EVC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein (DWF-1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.012819 (rank : 136) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.012639 (rank : 137) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.012409 (rank : 138) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.011980 (rank : 139) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CCD11_MOUSE
|
||||||
| NC score | 0.011961 (rank : 140) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.011951 (rank : 141) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.011469 (rank : 142) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
GRAM3_MOUSE
|
||||||
| NC score | 0.011387 (rank : 143) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PEM6 | Gene names | Gramd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRAM domain-containing protein 3. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.011385 (rank : 144) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.010309 (rank : 145) | θ value | 0.0961366 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
PARP4_HUMAN
|
||||||
| NC score | 0.009913 (rank : 146) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKK3, O75903, Q14682, Q9H1M6 | Gene names | PARP4, ADPRTL1, KIAA0177, PARPL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 4 (EC 2.4.2.30) (PARP-4) (Vault poly(ADP- ribose) polymerase) (VPARP) (193 kDa vault protein) (PARP- related/IalphaI-related H5/proline-rich) (PH5P). | |||||
|
TPM2_HUMAN
|
||||||
| NC score | 0.009789 (rank : 147) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TPM2_MOUSE
|
||||||
| NC score | 0.009707 (rank : 148) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.009369 (rank : 149) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.008724 (rank : 150) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.008323 (rank : 151) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MGRN1_HUMAN
|
||||||
| NC score | 0.008323 (rank : 152) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.007280 (rank : 153) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
CCD82_MOUSE
|
||||||
| NC score | 0.007266 (rank : 154) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PG04, Q3V462 | Gene names | Ccdc82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
KIF11_MOUSE
|
||||||
| NC score | 0.006855 (rank : 155) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6P9P6, Q9Z1J0 | Gene names | Kif11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.006422 (rank : 156) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
CK035_MOUSE
|
||||||
| NC score | 0.006333 (rank : 157) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q0VET5, Q9CW10 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf35 homolog. | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.005628 (rank : 158) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.004748 (rank : 159) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
ACTN1_MOUSE
|
||||||
| NC score | 0.004478 (rank : 160) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ERR3_HUMAN
|
||||||
| NC score | 0.004375 (rank : 161) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62508, O75454, O96021, Q68DA0, Q6P274, Q6PK28, Q6TS38, Q9R1F3, Q9UNJ4 | Gene names | ESRRG, ERR3, ERRG2, KIAA0832, NR3B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estrogen-related receptor gamma (Estrogen receptor-related protein 3) (ERR gamma-2). | |||||
|
ERR3_MOUSE
|
||||||
| NC score | 0.004375 (rank : 162) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62509, O75454, O96021, Q8CHC9, Q9R1F3 | Gene names | Esrrg, Err3, Kiaa0832, Nr3b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estrogen-related receptor gamma (Estrogen receptor-related protein 3). | |||||
|
ACTN1_HUMAN
|
||||||
| NC score | 0.004073 (rank : 163) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.003895 (rank : 164) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.003852 (rank : 165) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.003840 (rank : 166) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | -0.001084 (rank : 167) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | -0.001501 (rank : 168) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | -0.001818 (rank : 169) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
PDPK1_MOUSE
|
||||||
| NC score | -0.004591 (rank : 170) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2A0, Q9R1D8, Q9R215 | Gene names | Pdpk1, Pdk1 | |||
|
Domain Architecture |

|
|||||
| Description | 3-phosphoinositide-dependent protein kinase 1 (EC 2.7.11.1) (mPDK1). | |||||
|
PDPK1_HUMAN
|
||||||
| NC score | -0.004600 (rank : 171) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15530 | Gene names | PDPK1, PDK1 | |||
|
Domain Architecture |

|
|||||
| Description | 3-phosphoinositide-dependent protein kinase 1 (EC 2.7.11.1) (hPDK1). | |||||