Please be patient as the page loads
|
FANCM_MOUSE
|
||||||
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FANCM_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.964189 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 3) | NC score | 0.569715 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 4) | NC score | 0.555134 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 5) | NC score | 0.556426 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 6) | NC score | 0.554999 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 7) | NC score | 0.549865 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
ERCC4_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 8) | NC score | 0.281698 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92889, O00140, Q8TD83 | Gene names | ERCC4, ERCC11, XPF | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4) (DNA-repair protein complementing XP-F cells) (Xeroderma pigmentosum group F-complementing protein). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 9) | NC score | 0.549159 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 10) | NC score | 0.490041 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 11) | NC score | 0.487765 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 12) | NC score | 0.454804 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 13) | NC score | 0.459090 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
ERCC4_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 14) | NC score | 0.244332 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 15) | NC score | 0.457689 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 16) | NC score | 0.459614 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 17) | NC score | 0.451694 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 18) | NC score | 0.449249 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 19) | NC score | 0.449229 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 20) | NC score | 0.448608 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 21) | NC score | 0.433332 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 22) | NC score | 0.444921 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 23) | NC score | 0.445981 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 24) | NC score | 0.453489 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 25) | NC score | 0.444762 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 26) | NC score | 0.446332 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 27) | NC score | 0.447961 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 28) | NC score | 0.445975 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 29) | NC score | 0.445975 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 30) | NC score | 0.436383 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 31) | NC score | 0.429588 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 32) | NC score | 0.452191 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 33) | NC score | 0.235411 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 34) | NC score | 0.443991 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 35) | NC score | 0.444098 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 36) | NC score | 0.442491 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 37) | NC score | 0.442555 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 38) | NC score | 0.437378 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 39) | NC score | 0.443856 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 40) | NC score | 0.443856 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 41) | NC score | 0.436728 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 42) | NC score | 0.426125 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 43) | NC score | 0.425845 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 44) | NC score | 0.445310 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 45) | NC score | 0.444372 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 46) | NC score | 0.075268 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 47) | NC score | 0.420655 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 48) | NC score | 0.419907 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 49) | NC score | 0.433014 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 50) | NC score | 0.432998 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 51) | NC score | 0.423102 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 52) | NC score | 0.423774 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 53) | NC score | 0.428950 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 54) | NC score | 0.428911 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 55) | NC score | 0.421133 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 56) | NC score | 0.432459 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 57) | NC score | 0.432335 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 58) | NC score | 0.402175 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 59) | NC score | 0.422161 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 60) | NC score | 0.047220 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 61) | NC score | 0.067063 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 62) | NC score | 0.421075 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
U520_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 63) | NC score | 0.200565 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 64) | NC score | 0.402259 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 65) | NC score | 0.429135 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 66) | NC score | 0.424093 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 67) | NC score | 0.064693 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 68) | NC score | 0.425517 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 69) | NC score | 0.424162 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 70) | NC score | 0.415178 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 71) | NC score | 0.415135 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 72) | NC score | 0.416060 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
STAG1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 73) | NC score | 0.053707 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 74) | NC score | 0.414556 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 75) | NC score | 0.414765 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 76) | NC score | 0.414203 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 77) | NC score | 0.420822 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 78) | NC score | 0.022756 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 79) | NC score | 0.406289 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 80) | NC score | 0.228515 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 81) | NC score | 0.404540 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
STAG1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 82) | NC score | 0.052111 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D3E6, O08982 | Gene names | Stag1, Sa1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 83) | NC score | 0.043561 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.125558 (rank : 84) | NC score | 0.070402 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 0.125558 (rank : 85) | NC score | 0.403114 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.125558 (rank : 86) | NC score | 0.013536 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 0.163984 (rank : 87) | NC score | 0.411806 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 0.163984 (rank : 88) | NC score | 0.013641 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NRBF2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 89) | NC score | 0.060648 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCQ3, Q9DCG3 | Gene names | Nrbf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 0.21417 (rank : 90) | NC score | 0.403728 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
CO3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 91) | NC score | 0.030492 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01024 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment]. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 92) | NC score | 0.039312 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 93) | NC score | 0.025384 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
AQR_HUMAN
|
||||||
| θ value | 0.365318 (rank : 94) | NC score | 0.064866 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
AQR_MOUSE
|
||||||
| θ value | 0.365318 (rank : 95) | NC score | 0.067310 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
HXC12_HUMAN
|
||||||
| θ value | 0.47712 (rank : 96) | NC score | 0.014966 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31275, Q9BXJ6 | Gene names | HOXC12, HOX3F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C12 (Hox-3F). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 97) | NC score | 0.051176 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 98) | NC score | 0.039387 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 0.62314 (rank : 99) | NC score | 0.017935 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 100) | NC score | 0.275284 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 1.06291 (rank : 101) | NC score | 0.062880 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
HXC12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 102) | NC score | 0.013185 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K5B8 | Gene names | Hoxc12, Hox-3.8, Hoxc-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-C12 (Hox-3.8). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 103) | NC score | 0.030907 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 104) | NC score | 0.026724 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
CO3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 105) | NC score | 0.026071 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01027, Q61370 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor (HSE-MSF) [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3, isoform Short; Complement C3f fragment]. | |||||
|
GNPTA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 106) | NC score | 0.024246 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q69ZN6, Q3U3K6, Q3US34 | Gene names | Gnptab, Gnpta, Kiaa1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 107) | NC score | 0.017941 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PSTK_HUMAN
|
||||||
| θ value | 1.38821 (rank : 108) | NC score | 0.033424 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IV42, Q6ZSS9 | Gene names | PSTK, C10orf89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-seryl-tRNA(Ser/Sec) kinase (EC 2.7.1.-) (Phosphoseryl-tRNA(Ser/Sec) kinase). | |||||
|
RIOK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 109) | NC score | 0.028713 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
TCEA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 110) | NC score | 0.019278 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |
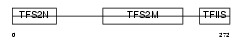
|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
DZIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 111) | NC score | 0.022169 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 112) | NC score | 0.036796 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
INADL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 113) | NC score | 0.009568 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 114) | NC score | 0.044574 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 115) | NC score | 0.028307 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 116) | NC score | 0.014294 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PHF22_HUMAN
|
||||||
| θ value | 1.81305 (rank : 117) | NC score | 0.031840 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
RAD54_HUMAN
|
||||||
| θ value | 1.81305 (rank : 118) | NC score | 0.050614 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
RAD54_MOUSE
|
||||||
| θ value | 1.81305 (rank : 119) | NC score | 0.051169 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 120) | NC score | 0.036264 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 121) | NC score | 0.198269 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
CENPC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 122) | NC score | 0.029310 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
ZFP91_MOUSE
|
||||||
| θ value | 2.36792 (rank : 123) | NC score | -0.000865 (rank : 189) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62511, Q8BPY3, Q8C2B4 | Gene names | Zfp91, Pzf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 (Zfp-91) (Zinc finger protein PZF) (Penta Zf protein). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 3.0926 (rank : 124) | NC score | 0.019137 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 125) | NC score | 0.009490 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
LN28B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 126) | NC score | 0.017309 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
MSPD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 127) | NC score | 0.023472 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NHP6, Q8N3H2, Q8NA83 | Gene names | MOSPD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 2. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 128) | NC score | 0.012736 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NMDE1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 129) | NC score | 0.010450 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
NRBF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 130) | NC score | 0.044121 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96F24, Q86UR2, Q96NP6, Q9H0S9, Q9H2I2 | Gene names | NRBF2, COPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2) (Comodulator of PPAR and RXR). | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 131) | NC score | 0.021221 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
STAG3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 132) | NC score | 0.038595 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJ98, Q8NDP3 | Gene names | STAG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
CI082_MOUSE
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.021412 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDY9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf82 homolog. | |||||
|
ICAL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 134) | NC score | 0.030375 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
K1718_HUMAN
|
||||||
| θ value | 4.03905 (rank : 135) | NC score | 0.012302 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 136) | NC score | 0.014987 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NMDE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 137) | NC score | 0.010079 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 138) | NC score | 0.029972 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
ZFP91_HUMAN
|
||||||
| θ value | 4.03905 (rank : 139) | NC score | -0.000821 (rank : 188) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.002529 (rank : 182) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
DPOD3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.027961 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
FRS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.011774 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
M3K5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | -0.004368 (rank : 191) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35099 | Gene names | Map3k5, Ask1, Mekk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.017528 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
OTC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 145) | NC score | 0.016877 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11725 | Gene names | Otc | |||
|
Domain Architecture |
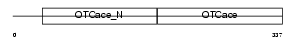
|
|||||
| Description | Ornithine carbamoyltransferase, mitochondrial precursor (EC 2.1.3.3) (OTCase) (Ornithine transcarbamylase). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 146) | NC score | 0.020703 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 147) | NC score | 0.016501 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
TIM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 148) | NC score | 0.014851 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNS1, O94802, Q86VM1, Q8IWH3 | Gene names | TIMELESS, TIM, TIM1, TIMELESS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Timeless homolog (hTIM). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 149) | NC score | 0.043422 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
VAPB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 150) | NC score | 0.016769 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95292, O95293, Q9P0H0 | Gene names | VAPB | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B/C (VAMP- associated protein B/C) (VAMP-B/VAMP-C) (VAP-B/VAP-C). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.017906 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ACM5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | -0.003157 (rank : 190) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08912, Q96RG7 | Gene names | CHRM5 | |||
|
Domain Architecture |
|
|||||
| Description | Muscarinic acetylcholine receptor M5. | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.008554 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.010180 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.010817 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.387134 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.010012 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.012665 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
NUAK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | -0.005187 (rank : 192) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 881 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q641K5, Q6I6D6, Q8CGE1 | Gene names | Nuak1, Kiaa0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1). | |||||
|
P55G_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.006535 (rank : 181) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
PAX3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.001086 (rank : 187) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-3. | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.042142 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TM131_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.020022 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
ARD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.001295 (rank : 186) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 621 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36406, Q9BZY4, Q9BZY5 | Gene names | TRIM23, ARD1, ARFD1, RNF46 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23) (RING finger protein 46). | |||||
|
ARD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.001703 (rank : 185) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGX0, Q8C2B6, Q8CDA4, Q8CDA7 | Gene names | Trim23, Arfd1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.016480 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.009489 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.007013 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
LRP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.001873 (rank : 183) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
MYPC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.001744 (rank : 184) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XKE0, Q8C109, Q8K2V0 | Gene names | Mybpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
NPM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.013620 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.016089 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.018545 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
SF3B3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.012300 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15393, Q9UJ29 | Gene names | SF3B3, KIAA0017, SAP130 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor 3B subunit 3 (Spliceosome-associated protein 130) (SAP 130) (SF3b130) (Pre-mRNA-splicing factor SF3b 130 kDa subunit) (STAF130). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.008571 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
THAP7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.011158 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |
|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.011504 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
BLM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.191827 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
BLM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.196929 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
CC026_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.070846 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
CHD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.053097 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.053444 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.382449 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.384397 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.375242 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.374835 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.184901 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.182738 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.236610 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.058700 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
SMCA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.050357 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
WRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.203050 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.964189 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.569715 (rank : 3) | θ value | 3.39556e-23 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.556426 (rank : 4) | θ value | 2.35696e-16 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.555134 (rank : 5) | θ value | 1.74391e-19 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.554999 (rank : 6) | θ value | 6.00763e-12 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.549865 (rank : 7) | θ value | 1.33837e-11 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.549159 (rank : 8) | θ value | 2.52405e-10 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.490041 (rank : 9) | θ value | 2.79066e-09 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.487765 (rank : 10) | θ value | 6.21693e-09 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.459614 (rank : 11) | θ value | 1.09739e-05 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.459090 (rank : 12) | θ value | 2.21117e-06 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.457689 (rank : 13) | θ value | 8.40245e-06 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.454804 (rank : 14) | θ value | 9.92553e-07 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.453489 (rank : 15) | θ value | 7.1131e-05 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.452191 (rank : 16) | θ value | 9.29e-05 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.451694 (rank : 17) | θ value | 1.43324e-05 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.449249 (rank : 18) | θ value | 1.43324e-05 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.449229 (rank : 19) | θ value | 1.43324e-05 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.448608 (rank : 20) | θ value | 1.87187e-05 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.447961 (rank : 21) | θ value | 7.1131e-05 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.446332 (rank : 22) | θ value | 7.1131e-05 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.445981 (rank : 23) | θ value | 4.1701e-05 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.445975 (rank : 24) | θ value | 7.1131e-05 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.445975 (rank : 25) | θ value | 7.1131e-05 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.445310 (rank : 26) | θ value | 0.00035302 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.444921 (rank : 27) | θ value | 4.1701e-05 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.444762 (rank : 28) | θ value | 7.1131e-05 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.444372 (rank : 29) | θ value | 0.000461057 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.444098 (rank : 30) | θ value | 0.000158464 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.443991 (rank : 31) | θ value | 0.000158464 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.443856 (rank : 32) | θ value | 0.00020696 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.443856 (rank : 33) | θ value | 0.00020696 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.442555 (rank : 34) | θ value | 0.00020696 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.442491 (rank : 35) | θ value | 0.000158464 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.437378 (rank : 36) | θ value | 0.00020696 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.436728 (rank : 37) | θ value | 0.000270298 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.436383 (rank : 38) | θ value | 9.29e-05 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.433332 (rank : 39) | θ value | 4.1701e-05 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.433014 (rank : 40) | θ value | 0.00298849 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.432998 (rank : 41) | θ value | 0.00298849 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.432459 (rank : 42) | θ value | 0.00509761 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.432335 (rank : 43) | θ value | 0.00509761 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.429588 (rank : 44) | θ value | 9.29e-05 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.429135 (rank : 45) | θ value | 0.0193708 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.428950 (rank : 46) | θ value | 0.00390308 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.428911 (rank : 47) | θ value | 0.00390308 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.426125 (rank : 48) | θ value | 0.000270298 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.425845 (rank : 49) | θ value | 0.000270298 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.425517 (rank : 50) | θ value | 0.0252991 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.424162 (rank : 51) | θ value | 0.0252991 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.424093 (rank : 52) | θ value | 0.0193708 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.423774 (rank : 53) | θ value | 0.00390308 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.423102 (rank : 54) | θ value | 0.00298849 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.422161 (rank : 55) | θ value | 0.00665767 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.421133 (rank : 56) | θ value | 0.00509761 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.421075 (rank : 57) | θ value | 0.00869519 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.420822 (rank : 58) | θ value | 0.0431538 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.420655 (rank : 59) | θ value | 0.000602161 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.419907 (rank : 60) | θ value | 0.00175202 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.416060 (rank : 61) | θ value | 0.0330416 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.415178 (rank : 62) | θ value | 0.0252991 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.415135 (rank : 63) | θ value | 0.0252991 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.414765 (rank : 64) | θ value | 0.0431538 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.414556 (rank : 65) | θ value | 0.0431538 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.414203 (rank : 66) | θ value | 0.0431538 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.411806 (rank : 67) | θ value | 0.163984 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.406289 (rank : 68) | θ value | 0.0563607 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.404540 (rank : 69) | θ value | 0.0736092 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.403728 (rank : 70) | θ value | 0.21417 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.403114 (rank : 71) | θ value | 0.125558 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.402259 (rank : 72) | θ value | 0.0148317 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.402175 (rank : 73) | θ value | 0.00665767 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.387134 (rank : 74) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.384397 (rank : 75) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.382449 (rank : 76) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.375242 (rank : 77) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.374835 (rank : 78) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
ERCC4_HUMAN
|
||||||
| NC score | 0.281698 (rank : 79) | θ value | 8.67504e-11 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92889, O00140, Q8TD83 | Gene names | ERCC4, ERCC11, XPF | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4) (DNA-repair protein complementing XP-F cells) (Xeroderma pigmentosum group F-complementing protein). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.275284 (rank : 80) | θ value | 0.813845 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
ERCC4_MOUSE
|
||||||
| NC score | 0.244332 (rank : 81) | θ value | 2.88788e-06 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.236610 (rank : 82) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.235411 (rank : 83) | θ value | 9.29e-05 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.228515 (rank : 84) | θ value | 0.0563607 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.203050 (rank : 85) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.200565 (rank : 86) | θ value | 0.00869519 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.198269 (rank : 87) | θ value | 1.81305 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.196929 (rank : 88) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.191827 (rank : 89) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.184901 (rank : 90) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.182738 (rank : 91) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.075268 (rank : 92) | θ value | 0.000602161 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.070846 (rank : 93) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.070402 (rank : 94) | θ value | 0.125558 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
AQR_MOUSE
|
||||||
| NC score | 0.067310 (rank : 95) | θ value | 0.365318 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.067063 (rank : 96) | θ value | 0.00869519 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
AQR_HUMAN
|
||||||
| NC score | 0.064866 (rank : 97) | θ value | 0.365318 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.064693 (rank : 98) | θ value | 0.0252991 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.062880 (rank : 99) | θ value | 1.06291 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
NRBF2_MOUSE
|
||||||
| NC score | 0.060648 (rank : 100) | θ value | 0.163984 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCQ3, Q9DCG3 | Gene names | Nrbf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.058700 (rank : 101) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
STAG1_HUMAN
|
||||||
| NC score | 0.053707 (rank : 102) | θ value | 0.0330416 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.053444 (rank : 103) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.053097 (rank : 104) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
STAG1_MOUSE
|
||||||
| NC score | 0.052111 (rank : 105) | θ value | 0.0736092 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D3E6, O08982 | Gene names | Stag1, Sa1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.051176 (rank : 106) | θ value | 0.62314 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
RAD54_MOUSE
|
||||||
| NC score | 0.051169 (rank : 107) | θ value | 1.81305 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70270, Q8BSR5, Q8C2C4, Q8K3D4 | Gene names | Rad54l, Rad54 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (mRAD54) (mHR54). | |||||
|
RAD54_HUMAN
|
||||||
| NC score | 0.050614 (rank : 108) | θ value | 1.81305 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92698, Q6IUY3 | Gene names | RAD54L, RAD54A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair and recombination protein RAD54-like (EC 3.6.1.-) (RAD54 homolog) (hRAD54) (hHR54). | |||||
|
SMCA1_MOUSE
|
||||||
| NC score | 0.050357 (rank : 109) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PGB8, Q8BSS1, Q91Y58 | Gene names | Smarca1, Snf2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable global transcription activator SNF2L1 (EC 3.6.1.-) (Nucleosome remodeling factor subunit SNF2L) (ATP-dependent helicase SMARCA1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 1) (DNA-dependent ATPase SNF2L). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.047220 (rank : 110) | θ value | 0.00665767 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.044574 (rank : 111) | θ value | 1.81305 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NRBF2_HUMAN
|
||||||
| NC score | 0.044121 (rank : 112) | θ value | 3.0926 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96F24, Q86UR2, Q96NP6, Q9H0S9, Q9H2I2 | Gene names | NRBF2, COPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2) (Comodulator of PPAR and RXR). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.043561 (rank : 113) | θ value | 0.0961366 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.043422 (rank : 114) | θ value | 5.27518 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.042142 (rank : 115) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.039387 (rank : 116) | θ value | 0.62314 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.039312 (rank : 117) | θ value | 0.279714 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
STAG3_HUMAN
|
||||||
| NC score | 0.038595 (rank : 118) | θ value | 3.0926 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJ98, Q8NDP3 | Gene names | STAG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-3 (Stromal antigen 3) (Stromalin 3) (SCC3 homolog 3). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.036796 (rank : 119) | θ value | 1.81305 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.036264 (rank : 120) | θ value | 1.81305 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
PSTK_HUMAN
|
||||||
| NC score | 0.033424 (rank : 121) | θ value | 1.38821 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IV42, Q6ZSS9 | Gene names | PSTK, C10orf89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-seryl-tRNA(Ser/Sec) kinase (EC 2.7.1.-) (Phosphoseryl-tRNA(Ser/Sec) kinase). | |||||
|
PHF22_HUMAN
|
||||||
| NC score | 0.031840 (rank : 122) | θ value | 1.81305 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.030907 (rank : 123) | θ value | 1.06291 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
CO3_HUMAN
|
||||||
| NC score | 0.030492 (rank : 124) | θ value | 0.279714 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01024 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3f fragment]. | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.030375 (rank : 125) | θ value | 4.03905 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.029972 (rank : 126) | θ value | 4.03905 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
CENPC_HUMAN
|
||||||
| NC score | 0.029310 (rank : 127) | θ value | 2.36792 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.028713 (rank : 128) | θ value | 1.38821 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.028307 (rank : 129) | θ value | 1.81305 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
DPOD3_HUMAN
|
||||||
| NC score | 0.027961 (rank : 130) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.026724 (rank : 131) | θ value | 1.06291 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
CO3_MOUSE
|
||||||
| NC score | 0.026071 (rank : 132) | θ value | 1.38821 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01027, Q61370 | Gene names | C3 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C3 precursor (HSE-MSF) [Contains: Complement C3 beta chain; Complement C3 alpha chain; C3a anaphylatoxin; Complement C3b alpha' chain; Complement C3c fragment; Complement C3dg fragment; Complement C3g fragment; Complement C3d fragment; Complement C3, isoform Short; Complement C3f fragment]. | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.025384 (rank : 133) | θ value | 0.279714 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
GNPTA_MOUSE
|
||||||
| NC score | 0.024246 (rank : 134) | θ value | 1.38821 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q69ZN6, Q3U3K6, Q3US34 | Gene names | Gnptab, Gnpta, Kiaa1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
MSPD2_HUMAN
|
||||||
| NC score | 0.023472 (rank : 135) | θ value | 3.0926 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NHP6, Q8N3H2, Q8NA83 | Gene names | MOSPD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 2. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.022756 (rank : 136) | θ value | 0.0431538 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DZIP1_HUMAN
|
||||||
| NC score | 0.022169 (rank : 137) | θ value | 1.81305 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86YF9, Q8WY45, Q8WY46, Q9UGA5, Q9Y2K0 | Gene names | DZIP1, DZIP, DZIP2, KIAA0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1/2). | |||||
|
CI082_MOUSE
|
||||||
| NC score | 0.021412 (rank : 138) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDY9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf82 homolog. | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.021221 (rank : 139) | θ value | 3.0926 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.020703 (rank : 140) | θ value | 5.27518 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
TM131_MOUSE
|
||||||
| NC score | 0.020022 (rank : 141) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
TCEA1_HUMAN
|
||||||
| NC score | 0.019278 (rank : 142) | θ value | 1.38821 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.019137 (rank : 143) | θ value | 3.0926 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.018545 (rank : 144) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.017941 (rank : 145) | θ value | 1.38821 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.017935 (rank : 146) | θ value | 0.62314 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.017906 (rank : 147) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.017528 (rank : 148) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LN28B_MOUSE
|
||||||
| NC score | 0.017309 (rank : 149) | θ value | 3.0926 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
OTC_MOUSE
|
||||||
| NC score | 0.016877 (rank : 150) | θ value | 5.27518 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11725 | Gene names | Otc | |||
|
Domain Architecture |

|
|||||
| Description | Ornithine carbamoyltransferase, mitochondrial precursor (EC 2.1.3.3) (OTCase) (Ornithine transcarbamylase). | |||||
|
VAPB_HUMAN
|
||||||
| NC score | 0.016769 (rank : 151) | θ value | 5.27518 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95292, O95293, Q9P0H0 | Gene names | VAPB | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B/C (VAMP- associated protein B/C) (VAMP-B/VAMP-C) (VAP-B/VAP-C). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.016501 (rank : 152) | θ value | 5.27518 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.016480 (rank : 153) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.016089 (rank : 154) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.014987 (rank : 155) | θ value | 4.03905 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
HXC12_HUMAN
|
||||||
| NC score | 0.014966 (rank : 156) | θ value | 0.47712 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31275, Q9BXJ6 | Gene names | HOXC12, HOX3F | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C12 (Hox-3F). | |||||
|
TIM_HUMAN
|
||||||
| NC score | 0.014851 (rank : 157) | θ value | 5.27518 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNS1, O94802, Q86VM1, Q8IWH3 | Gene names | TIMELESS, TIM, TIM1, TIMELESS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Timeless homolog (hTIM). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.014294 (rank : 158) | θ value | 1.81305 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.013641 (rank : 159) | θ value | 0.163984 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NPM_MOUSE
|
||||||
| NC score | 0.013620 (rank : 160) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.013536 (rank : 161) | θ value | 0.125558 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
HXC12_MOUSE
|
||||||
| NC score | 0.013185 (rank : 162) | θ value | 1.06291 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K5B8 | Gene names | Hoxc12, Hox-3.8, Hoxc-12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-C12 (Hox-3.8). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.012736 (rank : 163) | θ value | 3.0926 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.012665 (rank : 164) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.012302 (rank : 165) | θ value | 4.03905 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
SF3B3_HUMAN
|
||||||
| NC score | 0.012300 (rank : 166) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15393, Q9UJ29 | Gene names | SF3B3, KIAA0017, SAP130 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 3 (Spliceosome-associated protein 130) (SAP 130) (SF3b130) (Pre-mRNA-splicing factor SF3b 130 kDa subunit) (STAF130). | |||||
|
FRS2_HUMAN
|
||||||
| NC score | 0.011774 (rank : 167) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WU20, O43558, Q7LDQ6 | Gene names | FRS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT). | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.011504 (rank : 168) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
THAP7_HUMAN
|
||||||
| NC score | 0.011158 (rank : 169) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BT49 | Gene names | THAP7 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 7. | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.010817 (rank : 170) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
NMDE1_MOUSE
|
||||||
| NC score | 0.010450 (rank : 171) | θ value | 3.0926 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35436 | Gene names | Grin2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.010180 (rank : 172) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.010079 (rank : 173) | θ value | 4.03905 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.010012 (rank : 174) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.009568 (rank : 175) | θ value | 1.81305 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.009490 (rank : 176) | θ value | 3.0926 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.009489 (rank : 177) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.008571 (rank : 178) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.008554 (rank : 179) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.007013 (rank : 180) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.006535 (rank : 181) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.002529 (rank : 182) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
LRP5_HUMAN
|
||||||
| NC score | 0.001873 (rank : 183) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
MYPC2_MOUSE
|
||||||
| NC score | 0.001744 (rank : 184) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XKE0, Q8C109, Q8K2V0 | Gene names | Mybpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
ARD1_MOUSE
|
||||||
| NC score | 0.001703 (rank : 185) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGX0, Q8C2B6, Q8CDA4, Q8CDA7 | Gene names | Trim23, Arfd1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23). | |||||
|
ARD1_HUMAN
|
||||||
| NC score | 0.001295 (rank : 186) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 621 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36406, Q9BZY4, Q9BZY5 | Gene names | TRIM23, ARD1, ARFD1, RNF46 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23) (RING finger protein 46). | |||||
|
PAX3_MOUSE
|
||||||
| NC score | 0.001086 (rank : 187) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3. | |||||
|
ZFP91_HUMAN
|
||||||
| NC score | -0.000821 (rank : 188) | θ value | 4.03905 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
ZFP91_MOUSE
|
||||||
| NC score | -0.000865 (rank : 189) | θ value | 2.36792 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62511, Q8BPY3, Q8C2B4 | Gene names | Zfp91, Pzf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 (Zfp-91) (Zinc finger protein PZF) (Penta Zf protein). | |||||
|
ACM5_HUMAN
|
||||||
| NC score | -0.003157 (rank : 190) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08912, Q96RG7 | Gene names | CHRM5 | |||
|
Domain Architecture |

|
|||||
| Description | Muscarinic acetylcholine receptor M5. | |||||
|
M3K5_MOUSE
|
||||||
| NC score | -0.004368 (rank : 191) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35099 | Gene names | Map3k5, Ask1, Mekk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 5 (EC 2.7.11.25) (MAPK/ERK kinase kinase 5) (MEK kinase 5) (MEKK 5) (Apoptosis signal- regulating kinase 1) (ASK-1). | |||||
|
NUAK1_MOUSE
|
||||||
| NC score | -0.005187 (rank : 192) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 881 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q641K5, Q6I6D6, Q8CGE1 | Gene names | Nuak1, Kiaa0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1). | |||||