Please be patient as the page loads
|
DPOD3_HUMAN
|
||||||
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DPOD3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
DPOD3_MOUSE
|
||||||
| θ value | 3.47146e-177 (rank : 2) | NC score | 0.931224 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 3) | NC score | 0.044689 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 4) | NC score | 0.107124 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.113192 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.118101 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.130901 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.053540 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
NPM_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.074446 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.021082 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.027465 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
MCM3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.049436 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.030527 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.041541 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
HUCE1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.078043 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB85, Q8BHW3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebral protein 1 homolog. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.085274 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.058222 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.095304 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.046077 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TBX2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.025123 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.037970 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
UBXD2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.049246 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
INCE_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.033547 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.022592 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SEC62_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.086947 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.049278 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.089940 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.058460 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
ATE1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.056572 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2A5, Q9Z2A4 | Gene names | Ate1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.008074 (rank : 131) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.032550 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SEC62_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.088796 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99442, O00682, O00729 | Gene names | TLOC1, SEC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1) (hTP-1). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.028177 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
MCM3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.043324 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
|
MORC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.030965 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.063856 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.054175 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
GAB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.041431 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.042161 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.059584 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
KIF14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.014576 (rank : 124) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.065276 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
SENP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.024481 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0U3, Q86XC8 | Gene names | SENP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.044783 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.077681 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TBCD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.024482 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.023239 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.048068 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CT116_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.040452 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96HY6, Q9BW47 | Gene names | C20orf116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 precursor. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.085057 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
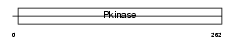
NEK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.006201 (rank : 133) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.055796 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.046199 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CT174_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.048230 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.014099 (rank : 125) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.043447 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.007047 (rank : 132) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
SSRP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.026643 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.069824 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
CAR10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.024933 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.027961 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
H15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.032680 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
H15_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.040077 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.052443 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
PRCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.042399 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.019983 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.044418 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
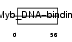
TERF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.028254 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |
|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TM131_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.044981 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
TSH3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.015189 (rank : 123) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.017274 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
UBIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.026390 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
XPO5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.020554 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAV4, Q5JTE6, Q96G48, Q96HN3, Q9BWM6, Q9BZV5, Q9H9M4, Q9NT89, Q9NW39, Q9ULC9 | Gene names | XPO5, KIAA1291, RANBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-5 (Exp5) (Ran-binding protein 21). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.048995 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.045927 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.027078 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
CT116_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.032897 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80WW9, Q9CT52 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 homolog precursor. | |||||
|
DEK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.048180 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ENL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.093925 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
K1210_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.050049 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.022513 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.049949 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.021391 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.044585 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
OSBL5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.011760 (rank : 129) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
P2RX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.013724 (rank : 126) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBL9, Q6V9R6, Q9NR37, Q9NR38, Q9UHD5, Q9UHD6, Q9UHD7, Q9Y637, Q9Y638 | Gene names | P2RX2, P2X2 | |||
|
Domain Architecture |
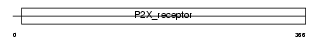
|
|||||
| Description | P2X purinoceptor 2 (ATP receptor) (P2X2) (Purinergic receptor). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.036441 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.034647 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PSIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.050209 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.020201 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.017726 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.041926 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.020489 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.002736 (rank : 134) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.015353 (rank : 122) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.013473 (rank : 127) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.018678 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.020007 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.019534 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.046355 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.030068 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.015601 (rank : 121) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.012734 (rank : 128) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.032164 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
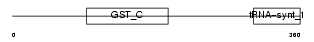
SYV_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.011501 (rank : 130) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |
|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.073921 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TDG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.038924 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13569, Q8IUZ6, Q8IZM3 | Gene names | TDG | |||
|
Domain Architecture |
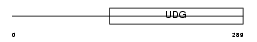
|
|||||
| Description | G/T mismatch-specific thymine DNA glycosylase (EC 3.2.2.-). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.068615 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.029049 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
AF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051038 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.052162 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.098909 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.065735 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.062713 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.055706 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.058304 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
H12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050364 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |
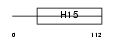
|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
H13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.053190 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.066711 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.068881 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.061495 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.090958 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.070334 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.076089 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.062718 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.060635 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.067666 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.066831 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.058180 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.050273 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.053005 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.057353 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.073355 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
VEZA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.065018 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
DPOD3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
DPOD3_MOUSE
|
||||||
| NC score | 0.931224 (rank : 2) | θ value | 3.47146e-177 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.130901 (rank : 3) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.118101 (rank : 4) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.113192 (rank : 5) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.107124 (rank : 6) | θ value | 0.0193708 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.098909 (rank : 7) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.095304 (rank : 8) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.093925 (rank : 9) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.090958 (rank : 10) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.089940 (rank : 11) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SEC62_HUMAN
|
||||||
| NC score | 0.088796 (rank : 12) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99442, O00682, O00729 | Gene names | TLOC1, SEC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1) (hTP-1). | |||||
|
SEC62_MOUSE
|
||||||
| NC score | 0.086947 (rank : 13) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BU14, Q6NX81 | Gene names | Tloc1, Sec62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translocation protein SEC62 (Translocation protein 1) (TP-1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.085274 (rank : 14) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.085057 (rank : 15) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
HUCE1_MOUSE
|
||||||
| NC score | 0.078043 (rank : 16) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DB85, Q8BHW3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebral protein 1 homolog. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.077681 (rank : 17) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.076089 (rank : 18) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
NPM_MOUSE
|
||||||
| NC score | 0.074446 (rank : 19) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.073921 (rank : 20) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.073355 (rank : 21) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.070334 (rank : 22) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.069824 (rank : 23) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.068881 (rank : 24) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.068615 (rank : 25) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.067666 (rank : 26) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.066831 (rank : 27) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.066711 (rank : 28) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.065735 (rank : 29) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.065276 (rank : 30) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
VEZA_HUMAN
|
||||||
| NC score | 0.065018 (rank : 31) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.063856 (rank : 32) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.062718 (rank : 33) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.062713 (rank : 34) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.061495 (rank : 35) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.060635 (rank : 36) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.059584 (rank : 37) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.058460 (rank : 38) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.058304 (rank : 39) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.058222 (rank : 40) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.058180 (rank : 41) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.057353 (rank : 42) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ATE1_MOUSE
|
||||||
| NC score | 0.056572 (rank : 43) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2A5, Q9Z2A4 | Gene names | Ate1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.055796 (rank : 44) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.055706 (rank : 45) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.054175 (rank : 46) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.053540 (rank : 47) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.053190 (rank : 48) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.053005 (rank : 49) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.052443 (rank : 50) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.052162 (rank : 51) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.051038 (rank : 52) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
H12_MOUSE
|
||||||
| NC score | 0.050364 (rank : 53) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.050273 (rank : 54) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
PSIP1_HUMAN
|
||||||
| NC score | 0.050209 (rank : 55) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75475, O00256, O95368, Q6P391, Q86YB9, Q9NZI3, Q9UER6 | Gene names | PSIP1, DFS70, LEDGF, PSIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (Transcriptional coactivator p75/p52) (Dense fine speckles 70 kDa protein) (DFS 70) (CLL-associated antigen KW-7). | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.050049 (rank : 56) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.049949 (rank : 57) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MCM3_HUMAN
|
||||||
| NC score | 0.049436 (rank : 58) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.049278 (rank : 59) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
UBXD2_HUMAN
|
||||||
| NC score | 0.049246 (rank : 60) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.048995 (rank : 61) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.048230 (rank : 62) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.048180 (rank : 63) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.048068 (rank : 64) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.046355 (rank : 65) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.046199 (rank : 66) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.046077 (rank : 67) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.045927 (rank : 68) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
TM131_MOUSE
|
||||||
| NC score | 0.044981 (rank : 69) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.044783 (rank : 70) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.044689 (rank : 71) | θ value | 0.00869519 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.044585 (rank : 72) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.044418 (rank : 73) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.043447 (rank : 74) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
MCM3_MOUSE
|
||||||
| NC score | 0.043324 (rank : 75) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
|
PRCC_HUMAN
|
||||||
| NC score | 0.042399 (rank : 76) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.042161 (rank : 77) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.041926 (rank : 78) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.041541 (rank : 79) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
GAB1_HUMAN
|
||||||
| NC score | 0.041431 (rank : 80) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13480, Q4W5G2, Q6P1W2 | Gene names | GAB1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
CT116_HUMAN
|
||||||
| NC score | 0.040452 (rank : 81) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96HY6, Q9BW47 | Gene names | C20orf116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 precursor. | |||||
|
H15_MOUSE
|
||||||
| NC score | 0.040077 (rank : 82) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
TDG_HUMAN
|
||||||
| NC score | 0.038924 (rank : 83) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13569, Q8IUZ6, Q8IZM3 | Gene names | TDG | |||
|
Domain Architecture |

|
|||||
| Description | G/T mismatch-specific thymine DNA glycosylase (EC 3.2.2.-). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.037970 (rank : 84) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.036441 (rank : 85) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.034647 (rank : 86) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.033547 (rank : 87) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CT116_MOUSE
|
||||||
| NC score | 0.032897 (rank : 88) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80WW9, Q9CT52 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 homolog precursor. | |||||
|
H15_HUMAN
|
||||||
| NC score | 0.032680 (rank : 89) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.032550 (rank : 90) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.032164 (rank : 91) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MORC2_HUMAN
|
||||||
| NC score | 0.030965 (rank : 92) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.030527 (rank : 93) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.030068 (rank : 94) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.029049 (rank : 95) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
TERF2_HUMAN
|
||||||
| NC score | 0.028254 (rank : 96) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15554 | Gene names | TERF2, TRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.028177 (rank : 97) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.027961 (rank : 98) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.027465 (rank : 99) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.027078 (rank : 100) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
SSRP1_HUMAN
|
||||||
| NC score | 0.026643 (rank : 101) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08945, Q5BJG8 | Gene names | SSRP1, FACT80 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (FACT 80 kDa subunit) (FACTp80) (Chromatin- specific transcription elongation factor 80 kDa subunit) (Structure- specific recognition protein 1) (hSSRP1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
UBIP1_HUMAN
|
||||||
| NC score | 0.026390 (rank : 102) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
TBX2_HUMAN
|
||||||
| NC score | 0.025123 (rank : 103) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
CAR10_HUMAN
|
||||||
| NC score | 0.024933 (rank : 104) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
TBCD1_MOUSE
|
||||||
| NC score | 0.024482 (rank : 105) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
SENP1_HUMAN
|
||||||
| NC score | 0.024481 (rank : 106) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0U3, Q86XC8 | Gene names | SENP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.023239 (rank : 107) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.022592 (rank : 108) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.022513 (rank : 109) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.021391 (rank : 110) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.021082 (rank : 111) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
XPO5_HUMAN
|
||||||
| NC score | 0.020554 (rank : 112) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAV4, Q5JTE6, Q96G48, Q96HN3, Q9BWM6, Q9BZV5, Q9H9M4, Q9NT89, Q9NW39, Q9ULC9 | Gene names | XPO5, KIAA1291, RANBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-5 (Exp5) (Ran-binding protein 21). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.020489 (rank : 113) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.020201 (rank : 114) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.020007 (rank : 115) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.019983 (rank : 116) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.019534 (rank : 117) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.018678 (rank : 118) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.017726 (rank : 119) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.017274 (rank : 120) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.015601 (rank : 121) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.015353 (rank : 122) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
TSH3_MOUSE
|
||||||
| NC score | 0.015189 (rank : 123) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
KIF14_HUMAN
|
||||||
| NC score | 0.014576 (rank : 124) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.014099 (rank : 125) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
P2RX2_HUMAN
|
||||||
| NC score | 0.013724 (rank : 126) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBL9, Q6V9R6, Q9NR37, Q9NR38, Q9UHD5, Q9UHD6, Q9UHD7, Q9Y637, Q9Y638 | Gene names | P2RX2, P2X2 | |||
|
Domain Architecture |
|
|||||
| Description | P2X purinoceptor 2 (ATP receptor) (P2X2) (Purinergic receptor). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.013473 (rank : 127) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.012734 (rank : 128) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
OSBL5_HUMAN
|
||||||
| NC score | 0.011760 (rank : 129) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
SYV_MOUSE
|
||||||
| NC score | 0.011501 (rank : 130) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1Q9, Q9QUN2 | Gene names | Vars, Bat6, G7a, Vars2 | |||
|
Domain Architecture |

|
|||||
| Description | Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA ligase) (ValRS) (Protein G7a). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | 0.008074 (rank : 131) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.007047 (rank : 132) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
NEK1_HUMAN
|
||||||
| NC score | 0.006201 (rank : 133) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.002736 (rank : 134) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||