Please be patient as the page loads
|
UBIP1_HUMAN
|
||||||
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TFCP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986821 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12800, Q12801, Q9UD77 | Gene names | TFCP2, LSF, SEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-globin transcription factor CP2 (Transcription factor LSF) (SAA3 enhancer factor). | |||||
|
TFCP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.986691 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ERA0, Q3UGJ4, Q8VC13 | Gene names | Tcfcp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-globin transcription factor CP2. | |||||
|
UBIP1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
UBIP1_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.994564 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
TF2L1_HUMAN
|
||||||
| θ value | 3.1398e-178 (rank : 5) | NC score | 0.985101 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZI6, Q4ZG43 | Gene names | TFCP2L1, CRTR1, LBP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor CP2-like protein 1 (CP2-related transcriptional repressor 1) (CRTR-1) (Transcription factor LBP-9). | |||||
|
TF2L1_MOUSE
|
||||||
| θ value | 1.78289e-173 (rank : 6) | NC score | 0.984751 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UNW5, Q99PF2 | Gene names | Tcfcp2l1, Crtr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor CP2-like protein 1 (CP2-related transcriptional repressor 1) (CRTR-1). | |||||
|
GRHL1_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 7) | NC score | 0.674480 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q921D9, Q8K5C1 | Gene names | Grhl1, Mgr, Tcfcp2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 1 homolog (Transcription factor CP2-like 2) (Transcription factor LBP-32). | |||||
|
GRHL1_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 8) | NC score | 0.667455 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZI5, Q53T93, Q6NWN7, Q6NWN8, Q6NWN9, Q8NI33 | Gene names | GRHL1, LBP32, MGR, TFCP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 1 homolog (Transcription factor CP2-like 2) (Transcription factor LBP-32) (NH32) (Mammalian grainyhead). | |||||
|
GRHL2_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 9) | NC score | 0.654228 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ISB3, Q6NT03, Q9H8B8 | Gene names | GRHL2, BOM, TFCP2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 2 homolog (Brother of mammalian grainyhead) (Transcription factor CP2-like 3). | |||||
|
GRHL2_MOUSE
|
||||||
| θ value | 7.82807e-20 (rank : 10) | NC score | 0.656590 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K5C0, Q80UZ5 | Gene names | Grhl2, Bom, Tcfcp2l3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 2 homolog (Brother of mammalian grainyhead) (Transcription factor CP2-like 3). | |||||
|
GRHL3_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 11) | NC score | 0.663162 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TE85, Q5TH78, Q86Y06, Q8N407 | Gene names | GRHL3, SOM, TFCP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 3 homolog (Sister of mammalian grainyhead) (Transcription factor CP2-like 4). | |||||
|
GRHL3_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 12) | NC score | 0.657205 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5FWH3 | Gene names | Grhl3, Tfcp2l4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 3 homolog (Transcription factor CP2-like 4). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.011415 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.044912 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.033901 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
ES8L1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.053171 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R5F8, Q8R0D6, Q9D2M6 | Gene names | Eps8l1, Eps8r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.025365 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.020132 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
PHX2A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.015505 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62066 | Gene names | Phox2a, Arix, Phox2, Pmx2, Pmx2a | |||
|
Domain Architecture |
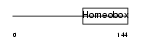
|
|||||
| Description | Paired mesoderm homeobox protein 2A (Paired-like homeobox 2A) (PHOX2A homeodomain protein) (Aristaless homeobox protein homolog). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.007284 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
K0460_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.041577 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
ADDA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.035290 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.023497 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
DPOD3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.047798 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
SMS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.043776 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86VZ5 | Gene names | TMEM23, MOB | |||
|
Domain Architecture |
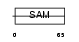
|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
SMS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.043747 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCQ6, Q8C464, Q8C583, Q8C652 | Gene names | Tmem23 | |||
|
Domain Architecture |
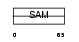
|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.020695 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.021705 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
ELL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.021732 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00472 | Gene names | ELL2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL2. | |||||
|
ES8L2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.047654 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H6S3, Q53GM8, Q8WYW7, Q96K06, Q9H6K9 | Gene names | EPS8L2, EPS8R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.002912 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.037105 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PHX2A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.009632 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14813, Q8IVZ2 | Gene names | PHOX2A, ARIX, PMX2A | |||
|
Domain Architecture |
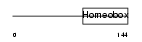
|
|||||
| Description | Paired mesoderm homeobox protein 2A (Paired-like homeobox 2A) (Aristaless homeobox protein homolog) (ARIX1 homeodomain protein). | |||||
|
SN1L2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.002498 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
APC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.025579 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.026710 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
BTBDA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.026070 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80X66 | Gene names | Btbd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 10. | |||||
|
CLPT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.030348 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O96005, Q53ET6, Q9BSS5 | Gene names | CLPTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleft lip and palate transmembrane protein 1. | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.016021 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.025289 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
ES8L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.031367 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99K30, Q91VT7 | Gene names | Eps8l2, Eps8r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
HSF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.012363 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03933, Q9H445 | Gene names | HSF2, HSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.009191 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
PPIL4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.024579 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.012154 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
GRPE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.024832 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LP6, Q3TSZ3 | Gene names | Grpel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GrpE protein homolog 1, mitochondrial precursor (Mt-GrpE#1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.012268 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.002772 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.018494 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.023600 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.009325 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
UBP2L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.037468 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
UBQL4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.017082 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRR5, Q9BR98, Q9UHX4 | Gene names | UBQLN4, C1orf6, UBIN | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.018752 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
BAG4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.018769 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CI61, Q3TRL9, Q91VT5, Q9CWG2 | Gene names | Bag4, Sodd | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
BTBDA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.022855 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BSF8, Q86WG1 | Gene names | BTBD10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 10. | |||||
|
DAAM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.011954 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DPOD3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.026390 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.014449 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.015983 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.015977 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.013930 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ZN467_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | -0.001769 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z7K2 | Gene names | ZNF467 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 467. | |||||
|
CCDC9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.020804 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
PININ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.009242 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
SPESP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.029672 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UW49, Q8NG22, Q8WVH8 | Gene names | SPESP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm equatorial segment protein 1 precursor (SP-ESP) (Equatorial segment protein) (ESP) (Glycosylated 38 kDa sperm protein C-7/8). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007479 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.015285 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
B4GN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.010916 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.006722 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.008900 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
DNM3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.011012 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.006475 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
IL18R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.004442 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61098 | Gene names | Il18r1 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-18 receptor 1 precursor (IL1 receptor-related protein) (IL-1Rrp). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.011888 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MST4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | -0.000006 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99JT2 | Gene names | Mst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MST4 (EC 2.7.11.1) (STE20-like kinase MST4) (MST-4) (Mammalian STE20-like protein kinase 4). | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.014244 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.009542 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PAPOA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.008795 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
SARM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.013895 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.003977 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.005255 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
UBP2L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.030839 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |
|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
UBIP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
UBIP1_MOUSE
|
||||||
| NC score | 0.994564 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
TFCP2_HUMAN
|
||||||
| NC score | 0.986821 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12800, Q12801, Q9UD77 | Gene names | TFCP2, LSF, SEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-globin transcription factor CP2 (Transcription factor LSF) (SAA3 enhancer factor). | |||||
|
TFCP2_MOUSE
|
||||||
| NC score | 0.986691 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ERA0, Q3UGJ4, Q8VC13 | Gene names | Tcfcp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-globin transcription factor CP2. | |||||
|
TF2L1_HUMAN
|
||||||
| NC score | 0.985101 (rank : 5) | θ value | 3.1398e-178 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NZI6, Q4ZG43 | Gene names | TFCP2L1, CRTR1, LBP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor CP2-like protein 1 (CP2-related transcriptional repressor 1) (CRTR-1) (Transcription factor LBP-9). | |||||
|
TF2L1_MOUSE
|
||||||
| NC score | 0.984751 (rank : 6) | θ value | 1.78289e-173 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UNW5, Q99PF2 | Gene names | Tcfcp2l1, Crtr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor CP2-like protein 1 (CP2-related transcriptional repressor 1) (CRTR-1). | |||||
|
GRHL1_MOUSE
|
||||||
| NC score | 0.674480 (rank : 7) | θ value | 1.6852e-22 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q921D9, Q8K5C1 | Gene names | Grhl1, Mgr, Tcfcp2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 1 homolog (Transcription factor CP2-like 2) (Transcription factor LBP-32). | |||||
|
GRHL1_HUMAN
|
||||||
| NC score | 0.667455 (rank : 8) | θ value | 1.86321e-21 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZI5, Q53T93, Q6NWN7, Q6NWN8, Q6NWN9, Q8NI33 | Gene names | GRHL1, LBP32, MGR, TFCP2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 1 homolog (Transcription factor CP2-like 2) (Transcription factor LBP-32) (NH32) (Mammalian grainyhead). | |||||
|
GRHL3_HUMAN
|
||||||
| NC score | 0.663162 (rank : 9) | θ value | 7.82807e-20 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TE85, Q5TH78, Q86Y06, Q8N407 | Gene names | GRHL3, SOM, TFCP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 3 homolog (Sister of mammalian grainyhead) (Transcription factor CP2-like 4). | |||||
|
GRHL3_MOUSE
|
||||||
| NC score | 0.657205 (rank : 10) | θ value | 5.07402e-19 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5FWH3 | Gene names | Grhl3, Tfcp2l4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 3 homolog (Transcription factor CP2-like 4). | |||||
|
GRHL2_MOUSE
|
||||||
| NC score | 0.656590 (rank : 11) | θ value | 7.82807e-20 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K5C0, Q80UZ5 | Gene names | Grhl2, Bom, Tcfcp2l3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 2 homolog (Brother of mammalian grainyhead) (Transcription factor CP2-like 3). | |||||
|
GRHL2_HUMAN
|
||||||
| NC score | 0.654228 (rank : 12) | θ value | 4.58923e-20 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ISB3, Q6NT03, Q9H8B8 | Gene names | GRHL2, BOM, TFCP2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Grainyhead-like protein 2 homolog (Brother of mammalian grainyhead) (Transcription factor CP2-like 3). | |||||
|
ES8L1_MOUSE
|
||||||
| NC score | 0.053171 (rank : 13) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R5F8, Q8R0D6, Q9D2M6 | Gene names | Eps8l1, Eps8r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
DPOD3_MOUSE
|
||||||
| NC score | 0.047798 (rank : 14) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
ES8L2_HUMAN
|
||||||
| NC score | 0.047654 (rank : 15) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H6S3, Q53GM8, Q8WYW7, Q96K06, Q9H6K9 | Gene names | EPS8L2, EPS8R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.044912 (rank : 16) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SMS1_HUMAN
|
||||||
| NC score | 0.043776 (rank : 17) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86VZ5 | Gene names | TMEM23, MOB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
SMS1_MOUSE
|
||||||
| NC score | 0.043747 (rank : 18) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCQ6, Q8C464, Q8C583, Q8C652 | Gene names | Tmem23 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine:ceramide cholinephosphotransferase 1 (EC 2.7.-.-) (Transmembrane protein 23) (Sphingomyelin synthase 1) (Mob protein). | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.041577 (rank : 19) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
UBP2L_MOUSE
|
||||||
| NC score | 0.037468 (rank : 20) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80X50, Q8BIT6, Q8BIW4, Q8BJ01, Q8CIG7, Q8K102, Q9CRT6 | Gene names | Ubap2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-associated protein 2-like. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.037105 (rank : 21) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
ADDA_HUMAN
|
||||||
| NC score | 0.035290 (rank : 22) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.033901 (rank : 23) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
ES8L2_MOUSE
|
||||||
| NC score | 0.031367 (rank : 24) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99K30, Q91VT7 | Gene names | Eps8l2, Eps8r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
UBP2L_HUMAN
|
||||||
| NC score | 0.030839 (rank : 25) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14157, Q5VU75, Q5VU76, Q9BTU3, Q9UGL5 | Gene names | UBAP2L, KIAA0144, NICE4 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-associated protein 2-like (Protein NICE-4). | |||||
|
CLPT1_HUMAN
|
||||||
| NC score | 0.030348 (rank : 26) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O96005, Q53ET6, Q9BSS5 | Gene names | CLPTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleft lip and palate transmembrane protein 1. | |||||
|
SPESP_HUMAN
|
||||||
| NC score | 0.029672 (rank : 27) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UW49, Q8NG22, Q8WVH8 | Gene names | SPESP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm equatorial segment protein 1 precursor (SP-ESP) (Equatorial segment protein) (ESP) (Glycosylated 38 kDa sperm protein C-7/8). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.026710 (rank : 28) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
DPOD3_HUMAN
|
||||||
| NC score | 0.026390 (rank : 29) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
BTBDA_MOUSE
|
||||||
| NC score | 0.026070 (rank : 30) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80X66 | Gene names | Btbd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 10. | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.025579 (rank : 31) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.025365 (rank : 32) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.025289 (rank : 33) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
GRPE1_MOUSE
|
||||||
| NC score | 0.024832 (rank : 34) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LP6, Q3TSZ3 | Gene names | Grpel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GrpE protein homolog 1, mitochondrial precursor (Mt-GrpE#1). | |||||
|
PPIL4_MOUSE
|
||||||
| NC score | 0.024579 (rank : 35) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CXG3, Q3TJX1, Q68FC7, Q9CT22 | Gene names | Ppil4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.023600 (rank : 36) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.023497 (rank : 37) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
BTBDA_HUMAN
|
||||||
| NC score | 0.022855 (rank : 38) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BSF8, Q86WG1 | Gene names | BTBD10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 10. | |||||
|
ELL2_HUMAN
|
||||||
| NC score | 0.021732 (rank : 39) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00472 | Gene names | ELL2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL2. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.021705 (rank : 40) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CCDC9_HUMAN
|
||||||
| NC score | 0.020804 (rank : 41) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3X0 | Gene names | CCDC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.020695 (rank : 42) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.020132 (rank : 43) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
BAG4_MOUSE
|
||||||
| NC score | 0.018769 (rank : 44) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CI61, Q3TRL9, Q91VT5, Q9CWG2 | Gene names | Bag4, Sodd | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.018752 (rank : 45) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.018494 (rank : 46) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
UBQL4_HUMAN
|
||||||
| NC score | 0.017082 (rank : 47) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRR5, Q9BR98, Q9UHX4 | Gene names | UBQLN4, C1orf6, UBIN | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.016021 (rank : 48) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.015983 (rank : 49) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.015977 (rank : 50) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
PHX2A_MOUSE
|
||||||
| NC score | 0.015505 (rank : 51) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62066 | Gene names | Phox2a, Arix, Phox2, Pmx2, Pmx2a | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2A (Paired-like homeobox 2A) (PHOX2A homeodomain protein) (Aristaless homeobox protein homolog). | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.015285 (rank : 52) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.014449 (rank : 53) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.014244 (rank : 54) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.013930 (rank : 55) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SARM1_HUMAN
|
||||||
| NC score | 0.013895 (rank : 56) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
HSF2_HUMAN
|
||||||
| NC score | 0.012363 (rank : 57) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03933, Q9H445 | Gene names | HSF2, HSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.012268 (rank : 58) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.012154 (rank : 59) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DAAM1_HUMAN
|
||||||
| NC score | 0.011954 (rank : 60) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.011888 (rank : 61) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.011415 (rank : 62) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
DNM3B_HUMAN
|
||||||
| NC score | 0.011012 (rank : 63) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBC3, Q9UBD4, Q9UJQ5, Q9UKA6, Q9UNE5, Q9Y5R9, Q9Y5S0 | Gene names | DNMT3B | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3B (EC 2.1.1.37) (Dnmt3b) (DNA methyltransferase HsaIIIB) (DNA MTase HsaIIIB) (M.HsaIIIB). | |||||
|
B4GN3_HUMAN
|
||||||
| NC score | 0.010916 (rank : 64) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6L9W6, Q6ZNC1, Q8N7T6 | Gene names | B4GALNT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
PHX2A_HUMAN
|
||||||
| NC score | 0.009632 (rank : 65) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14813, Q8IVZ2 | Gene names | PHOX2A, ARIX, PMX2A | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2A (Paired-like homeobox 2A) (Aristaless homeobox protein homolog) (ARIX1 homeodomain protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.009542 (rank : 66) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.009325 (rank : 67) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.009242 (rank : 68) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.009191 (rank : 69) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.008900 (rank : 70) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
PAPOA_MOUSE
|
||||||
| NC score | 0.008795 (rank : 71) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.007479 (rank : 72) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.007284 (rank : 73) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.006722 (rank : 74) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.006475 (rank : 75) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.005255 (rank : 76) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
IL18R_MOUSE
|
||||||
| NC score | 0.004442 (rank : 77) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61098 | Gene names | Il18r1 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-18 receptor 1 precursor (IL1 receptor-related protein) (IL-1Rrp). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.003977 (rank : 78) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.002912 (rank : 79) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.002772 (rank : 80) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
SN1L2_MOUSE
|
||||||
| NC score | 0.002498 (rank : 81) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
MST4_MOUSE
|
||||||
| NC score | -0.000006 (rank : 82) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99JT2 | Gene names | Mst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MST4 (EC 2.7.11.1) (STE20-like kinase MST4) (MST-4) (Mammalian STE20-like protein kinase 4). | |||||
|
ZN467_HUMAN
|
||||||
| NC score | -0.001769 (rank : 83) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z7K2 | Gene names | ZNF467 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 467. | |||||