Please be patient as the page loads
|
ADDA_HUMAN
|
||||||
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ADDA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.966513 (rank : 2) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDG_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.959876 (rank : 4) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UEY8, O43243, Q92773, Q9UEY7 | Gene names | ADD3, ADDL | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDG_MOUSE
|
||||||
| θ value | 2.93876e-176 (rank : 4) | NC score | 0.963604 (rank : 3) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYB5, Q9D2M5, Q9QYB6 | Gene names | Add3, Addl | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 3.36238e-172 (rank : 5) | NC score | 0.849049 (rank : 6) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 7.25522e-167 (rank : 6) | NC score | 0.937028 (rank : 5) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 7) | NC score | 0.063237 (rank : 21) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 8) | NC score | 0.070965 (rank : 13) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 9) | NC score | 0.076491 (rank : 10) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 10) | NC score | 0.060390 (rank : 27) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 11) | NC score | 0.056001 (rank : 36) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.072127 (rank : 11) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.071770 (rank : 12) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.025290 (rank : 81) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.063017 (rank : 22) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.024346 (rank : 83) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.053581 (rank : 41) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.035651 (rank : 65) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.029490 (rank : 71) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
STK10_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.006515 (rank : 126) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.013431 (rank : 106) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.069732 (rank : 16) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.046612 (rank : 51) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.067956 (rank : 17) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
UBIP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.037529 (rank : 62) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.069957 (rank : 15) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.053652 (rank : 40) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CSPG5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.038261 (rank : 60) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
RAD18_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.052184 (rank : 44) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.045908 (rank : 53) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
IQGA2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.027867 (rank : 73) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.060605 (rank : 24) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.033583 (rank : 69) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
UBIP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.035290 (rank : 67) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
SLBP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.042577 (rank : 56) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97440 | Gene names | Slbp, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.013284 (rank : 107) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.036379 (rank : 64) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.039060 (rank : 59) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
ZN206_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.001053 (rank : 132) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.045173 (rank : 55) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
REV1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.035615 (rank : 66) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.035229 (rank : 68) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.037096 (rank : 63) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
TCEA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.027134 (rank : 76) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QVN7, O08667 | Gene names | Tcea2 | |||
|
Domain Architecture |
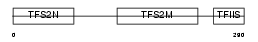
|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Protein S-II-T1) (Transcription elongation factor TFIIS.l). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.010634 (rank : 118) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
GLUD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.021232 (rank : 90) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIX8 | Gene names | Gluld1, Lgs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate--ammonia ligase domain-containing protein 1 (Lengsin) (Lens glutamine synthase-like). | |||||
|
LMD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.027349 (rank : 75) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
PTPRG_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.014162 (rank : 104) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.021601 (rank : 88) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TRI25_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.013014 (rank : 108) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61510 | Gene names | Trim25, Efp, Zfp147, Znf147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp). | |||||
|
4ET_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.018154 (rank : 97) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.042407 (rank : 57) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.027806 (rank : 74) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.002720 (rank : 130) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.032174 (rank : 70) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.021086 (rank : 91) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
FAIM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.013532 (rank : 105) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K097, Q3TY22, Q8K1F6, Q9D6K4 | Gene names | Faim2, Kiaa0950, Lfg | |||
|
Domain Architecture |
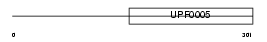
|
|||||
| Description | Fas apoptotic inhibitory molecule 2 (Lifeguard protein). | |||||
|
JARD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.025699 (rank : 79) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.026333 (rank : 77) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
K0494_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.014515 (rank : 102) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGQ6, Q3TQN6, Q8BJV7, Q8C0R1 | Gene names | Kiaa0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.019728 (rank : 94) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.026310 (rank : 78) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.022943 (rank : 84) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
SURF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.029401 (rank : 72) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15527, Q96CD1 | Gene names | SURF2 | |||
|
Domain Architecture |
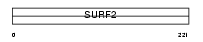
|
|||||
| Description | Surfeit locus protein 2 (Surf-2). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.012215 (rank : 111) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
ZN645_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.045507 (rank : 54) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.014393 (rank : 103) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
CAF1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.011206 (rank : 115) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D0N7 | Gene names | Chaf1b | |||
|
Domain Architecture |
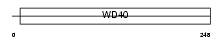
|
|||||
| Description | Chromatin assembly factor 1 subunit B (CAF-1 subunit B) (Chromatin assembly factor I p60 subunit) (CAF-I 60 kDa subunit) (CAF-Ip60). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.021358 (rank : 89) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.046034 (rank : 52) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.055348 (rank : 38) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.019726 (rank : 95) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
TRI56_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.008966 (rank : 122) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.011889 (rank : 112) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ZN688_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.000689 (rank : 133) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.022078 (rank : 86) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
BUB1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.012862 (rank : 109) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
C102B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.011853 (rank : 113) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CP2BJ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.003216 (rank : 129) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55071 | Gene names | Cyp2b19 | |||
|
Domain Architecture |
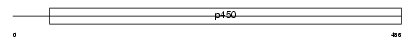
|
|||||
| Description | Cytochrome P450 2B19 (EC 1.14.14.1) (CYPIIB19). | |||||
|
FOXN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.010814 (rank : 117) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.015993 (rank : 99) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.020591 (rank : 93) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.011021 (rank : 116) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
MA1B1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.025537 (rank : 80) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.025003 (rank : 82) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PTPRG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.010057 (rank : 119) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.014954 (rank : 101) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.015495 (rank : 100) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
TNAP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.009629 (rank : 121) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.002445 (rank : 131) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.020789 (rank : 92) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
BRAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.007759 (rank : 124) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.011563 (rank : 114) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | -0.000292 (rank : 135) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
FATH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | -0.000116 (rank : 134) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.047539 (rank : 50) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.017251 (rank : 98) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.006266 (rank : 127) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.012498 (rank : 110) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.022658 (rank : 85) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SLY_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.006566 (rank : 125) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K352, Q3TBX0, Q9DBV2 | Gene names | Sly | |||
|
Domain Architecture |
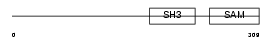
|
|||||
| Description | SH3 protein expressed in lymphocytes. | |||||
|
SRPK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.003314 (rank : 128) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.022002 (rank : 87) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.037774 (rank : 61) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
TJAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.008938 (rank : 123) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
TTF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.009860 (rank : 120) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15361, Q4VXF3, Q58EY2, Q6P5T5 | Gene names | TTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (RNA polymerase I termination factor). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.041506 (rank : 58) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.018881 (rank : 96) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.060495 (rank : 25) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.061790 (rank : 23) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.057023 (rank : 32) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CF010_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.057508 (rank : 31) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050843 (rank : 48) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.056989 (rank : 33) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.058527 (rank : 29) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.051709 (rank : 45) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.054264 (rank : 39) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.051571 (rank : 46) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.052243 (rank : 43) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.065088 (rank : 19) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.063817 (rank : 20) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.070584 (rank : 14) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.066871 (rank : 18) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.060392 (rank : 26) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.056614 (rank : 34) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.059568 (rank : 28) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050796 (rank : 49) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SNUT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.077611 (rank : 9) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
SNUT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.101334 (rank : 7) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K194, Q8BRA8, Q9CSV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.056557 (rank : 35) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.051005 (rank : 47) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.053456 (rank : 42) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.078363 (rank : 8) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.055619 (rank : 37) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.057624 (rank : 30) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ADDA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.966513 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDG_MOUSE
|
||||||
| NC score | 0.963604 (rank : 3) | θ value | 2.93876e-176 (rank : 4) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYB5, Q9D2M5, Q9QYB6 | Gene names | Add3, Addl | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDG_HUMAN
|
||||||
| NC score | 0.959876 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UEY8, O43243, Q92773, Q9UEY7 | Gene names | ADD3, ADDL | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.937028 (rank : 5) | θ value | 7.25522e-167 (rank : 6) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.849049 (rank : 6) | θ value | 3.36238e-172 (rank : 5) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
SNUT3_MOUSE
|
||||||
| NC score | 0.101334 (rank : 7) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K194, Q8BRA8, Q9CSV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.078363 (rank : 8) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
SNUT3_HUMAN
|
||||||
| NC score | 0.077611 (rank : 9) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.076491 (rank : 10) | θ value | 0.00509761 (rank : 9) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.072127 (rank : 11) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.071770 (rank : 12) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.070965 (rank : 13) | θ value | 0.00102713 (rank : 8) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.070584 (rank : 14) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.069957 (rank : 15) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.069732 (rank : 16) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.067956 (rank : 17) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.066871 (rank : 18) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.065088 (rank : 19) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.063817 (rank : 20) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.063237 (rank : 21) | θ value | 0.000786445 (rank : 7) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.063017 (rank : 22) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.061790 (rank : 23) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.060605 (rank : 24) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.060495 (rank : 25) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.060392 (rank : 26) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.060390 (rank : 27) | θ value | 0.00665767 (rank : 10) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.059568 (rank : 28) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.058527 (rank : 29) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.057624 (rank : 30) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.057508 (rank : 31) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.057023 (rank : 32) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.056989 (rank : 33) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.056614 (rank : 34) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.056557 (rank : 35) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.056001 (rank : 36) | θ value | 0.00869519 (rank : 11) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.055619 (rank : 37) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.055348 (rank : 38) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.054264 (rank : 39) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.053652 (rank : 40) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.053581 (rank : 41) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.053456 (rank : 42) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.052243 (rank : 43) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
RAD18_MOUSE
|
||||||
| NC score | 0.052184 (rank : 44) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.051709 (rank : 45) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.051571 (rank : 46) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.051005 (rank : 47) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.050843 (rank : 48) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.050796 (rank : 49) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.047539 (rank : 50) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.046612 (rank : 51) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.046034 (rank : 52) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.045908 (rank : 53) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ZN645_HUMAN
|
||||||
| NC score | 0.045507 (rank : 54) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.045173 (rank : 55) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SLBP_MOUSE
|
||||||
| NC score | 0.042577 (rank : 56) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97440 | Gene names | Slbp, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.042407 (rank : 57) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.041506 (rank : 58) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.039060 (rank : 59) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
CSPG5_HUMAN
|
||||||
| NC score | 0.038261 (rank : 60) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.037774 (rank : 61) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
UBIP1_MOUSE
|
||||||
| NC score | 0.037529 (rank : 62) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.037096 (rank : 63) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.036379 (rank : 64) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.035651 (rank : 65) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
REV1_MOUSE
|
||||||
| NC score | 0.035615 (rank : 66) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
UBIP1_HUMAN
|
||||||
| NC score | 0.035290 (rank : 67) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.035229 (rank : 68) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.033583 (rank : 69) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.032174 (rank : 70) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.029490 (rank : 71) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SURF2_HUMAN
|
||||||
| NC score | 0.029401 (rank : 72) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15527, Q96CD1 | Gene names | SURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 2 (Surf-2). | |||||
|
IQGA2_HUMAN
|
||||||
| NC score | 0.027867 (rank : 73) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13576 | Gene names | IQGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating-like protein IQGAP2. | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.027806 (rank : 74) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
LMD1_HUMAN
|
||||||
| NC score | 0.027349 (rank : 75) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
TCEA2_MOUSE
|
||||||
| NC score | 0.027134 (rank : 76) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QVN7, O08667 | Gene names | Tcea2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Protein S-II-T1) (Transcription elongation factor TFIIS.l). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.026333 (rank : 77) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.026310 (rank : 78) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.025699 (rank : 79) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
MA1B1_HUMAN
|
||||||
| NC score | 0.025537 (rank : 80) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.025290 (rank : 81) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.025003 (rank : 82) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.024346 (rank : 83) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.022943 (rank : 84) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.022658 (rank : 85) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.022078 (rank : 86) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.022002 (rank : 87) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.021601 (rank : 88) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.021358 (rank : 89) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
GLUD1_MOUSE
|
||||||
| NC score | 0.021232 (rank : 90) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIX8 | Gene names | Gluld1, Lgs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate--ammonia ligase domain-containing protein 1 (Lengsin) (Lens glutamine synthase-like). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.021086 (rank : 91) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.020789 (rank : 92) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.020591 (rank : 93) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.019728 (rank : 94) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.019726 (rank : 95) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.018881 (rank : 96) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
4ET_MOUSE
|
||||||
| NC score | 0.018154 (rank : 97) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.017251 (rank : 98) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.015993 (rank : 99) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.015495 (rank : 100) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.014954 (rank : 101) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
K0494_MOUSE
|
||||||
| NC score | 0.014515 (rank : 102) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGQ6, Q3TQN6, Q8BJV7, Q8C0R1 | Gene names | Kiaa0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.014393 (rank : 103) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
PTPRG_HUMAN
|
||||||
| NC score | 0.014162 (rank : 104) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23470, Q15623 | Gene names | PTPRG | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
FAIM2_MOUSE
|
||||||
| NC score | 0.013532 (rank : 105) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K097, Q3TY22, Q8K1F6, Q9D6K4 | Gene names | Faim2, Kiaa0950, Lfg | |||
|
Domain Architecture |

|
|||||
| Description | Fas apoptotic inhibitory molecule 2 (Lifeguard protein). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.013431 (rank : 106) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.013284 (rank : 107) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
TRI25_MOUSE
|
||||||
| NC score | 0.013014 (rank : 108) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61510 | Gene names | Trim25, Efp, Zfp147, Znf147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp). | |||||
|
BUB1B_MOUSE
|
||||||
| NC score | 0.012862 (rank : 109) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.012498 (rank : 110) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.012215 (rank : 111) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.011889 (rank : 112) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
C102B_HUMAN
|
||||||
| NC score | 0.011853 (rank : 113) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.011563 (rank : 114) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
CAF1B_MOUSE
|
||||||
| NC score | 0.011206 (rank : 115) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D0N7 | Gene names | Chaf1b | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin assembly factor 1 subunit B (CAF-1 subunit B) (Chromatin assembly factor I p60 subunit) (CAF-I 60 kDa subunit) (CAF-Ip60). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.011021 (rank : 116) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
FOXN1_MOUSE
|
||||||
| NC score | 0.010814 (rank : 117) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61575 | Gene names | Foxn1, Fkh19, Hfh11, Whn | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/forkhead homolog 11) (HFH-11). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.010634 (rank : 118) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
PTPRG_MOUSE
|
||||||
| NC score | 0.010057 (rank : 119) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05909 | Gene names | Ptprg | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase gamma precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase gamma) (R-PTP-gamma). | |||||
|
TTF1_HUMAN
|
||||||
| NC score | 0.009860 (rank : 120) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15361, Q4VXF3, Q58EY2, Q6P5T5 | Gene names | TTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (RNA polymerase I termination factor). | |||||
|
TNAP3_HUMAN
|
||||||
| NC score | 0.009629 (rank : 121) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.008966 (rank : 122) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
TJAP1_MOUSE
|
||||||
| NC score | 0.008938 (rank : 123) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
BRAP_HUMAN
|
||||||
| NC score | 0.007759 (rank : 124) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z569, O43238, O75341 | Gene names | BRAP, RNF52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BRCA1-associated protein (EC 6.3.2.-) (BRAP2) (Impedes mitogenic signal propagation) (IMP) (RING finger protein 52) (NY-REN-63 antigen). | |||||
|
SLY_MOUSE
|
||||||
| NC score | 0.006566 (rank : 125) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K352, Q3TBX0, Q9DBV2 | Gene names | Sly | |||
|
Domain Architecture |

|
|||||
| Description | SH3 protein expressed in lymphocytes. | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.006515 (rank : 126) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.006266 (rank : 127) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
SRPK1_MOUSE
|
||||||
| NC score | 0.003314 (rank : 128) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
CP2BJ_MOUSE
|
||||||
| NC score | 0.003216 (rank : 129) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55071 | Gene names | Cyp2b19 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2B19 (EC 1.14.14.1) (CYPIIB19). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.002720 (rank : 130) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.002445 (rank : 131) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ZN206_HUMAN
|
||||||
| NC score | 0.001053 (rank : 132) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96SZ4 | Gene names | ZNF206, ZSCAN10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 206 (Zinc finger and SCAN domain-containing protein 10). | |||||
|
ZN688_HUMAN
|
||||||
| NC score | 0.000689 (rank : 133) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
FATH_HUMAN
|
||||||
| NC score | -0.000116 (rank : 134) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | -0.000292 (rank : 135) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 108 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||