Please be patient as the page loads
|
SNUT3_HUMAN
|
||||||
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SNUT3_HUMAN
|
||||||
| θ value | 4.01107e-24 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
SNUT3_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 2) | NC score | 0.969763 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K194, Q8BRA8, Q9CSV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3. | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 3) | NC score | 0.248427 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 4) | NC score | 0.239808 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.165707 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RBM15_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.179035 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.096245 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.137294 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.094616 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.134045 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
RB15B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.127458 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.213721 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.073268 (rank : 77) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.074071 (rank : 76) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
DMN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.075387 (rank : 72) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.096982 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.140592 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.052577 (rank : 158) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.127731 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.129834 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TBX2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.035088 (rank : 184) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.049530 (rank : 175) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.053504 (rank : 151) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.065466 (rank : 98) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CENPT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.064039 (rank : 103) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.079807 (rank : 59) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.096550 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.014205 (rank : 189) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.081674 (rank : 57) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
ZO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.041192 (rank : 180) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.126121 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CA065_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.075111 (rank : 74) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.055865 (rank : 141) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.061724 (rank : 116) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.133288 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.179733 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.047066 (rank : 177) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.035484 (rank : 183) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.061708 (rank : 117) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SEPT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.020140 (rank : 187) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UH03, Q6IBZ6, Q8N3P3 | Gene names | SEPT3, SEP3 | |||
|
Domain Architecture |
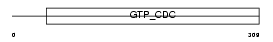
|
|||||
| Description | Neuronal-specific septin-3. | |||||
|
DCDC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.042980 (rank : 178) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.019771 (rank : 188) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
LRC27_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.047751 (rank : 176) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.100464 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.093882 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.010848 (rank : 190) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.027248 (rank : 185) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.041350 (rank : 179) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.022425 (rank : 186) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.051184 (rank : 168) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.038794 (rank : 181) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.037361 (rank : 182) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.098000 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ADDA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.077611 (rank : 64) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.084344 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.077307 (rank : 67) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UEY8, O43243, Q92773, Q9UEY7 | Gene names | ADD3, ADDL | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ADDG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.078405 (rank : 61) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QYB5, Q9D2M5, Q9QYB6 | Gene names | Add3, Addl | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.058596 (rank : 131) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.061033 (rank : 120) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.052641 (rank : 156) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.054172 (rank : 149) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.055206 (rank : 145) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.050487 (rank : 172) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.052135 (rank : 164) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.052500 (rank : 160) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.062027 (rank : 113) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.057738 (rank : 134) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.063287 (rank : 107) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.059743 (rank : 125) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.068225 (rank : 91) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.075325 (rank : 73) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.063849 (rank : 104) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.064883 (rank : 100) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
CF010_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.064235 (rank : 102) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CIR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.107107 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
CIR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.078014 (rank : 62) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CROP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.114867 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.113983 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
CS029_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.057734 (rank : 135) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
CTR9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.055245 (rank : 144) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.051097 (rank : 169) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.099632 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.096824 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DEK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.055626 (rank : 142) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DEK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.062035 (rank : 112) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.051898 (rank : 165) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.075644 (rank : 71) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.076277 (rank : 70) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.064593 (rank : 101) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.061651 (rank : 118) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.060367 (rank : 123) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.057320 (rank : 137) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
ENL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.058708 (rank : 130) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
FILA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.066656 (rank : 95) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
FIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.056590 (rank : 139) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.053062 (rank : 154) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.096038 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.083218 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.079246 (rank : 60) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.060908 (rank : 121) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.057708 (rank : 136) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
INCE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.062881 (rank : 109) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INCE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.065943 (rank : 97) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.066333 (rank : 96) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.063761 (rank : 105) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KI67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.050747 (rank : 170) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LC7L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.081109 (rank : 58) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
LC7L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.085075 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.067376 (rank : 93) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.058296 (rank : 132) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.062289 (rank : 111) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.068771 (rank : 88) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MINT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.072326 (rank : 79) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.064895 (rank : 99) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.070265 (rank : 84) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.143756 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.096883 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.050453 (rank : 173) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.089690 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.082295 (rank : 54) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.052867 (rank : 155) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PININ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.077792 (rank : 63) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PININ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.083064 (rank : 53) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
PRP40_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.077559 (rank : 65) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.083689 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.051289 (rank : 167) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.099701 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.113201 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.108336 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
REN3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.077197 (rank : 68) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BZI7, Q9H1J0 | Gene names | UPF3B, RENT3B, UPF3X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3B (Nonsense mRNA reducing factor 3B) (Up-frameshift suppressor 3 homolog B) (hUpf3B) (hUpf3p-X). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.070017 (rank : 86) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.068540 (rank : 89) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052504 (rank : 159) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |
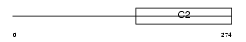
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
RIMS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.052457 (rank : 162) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U57 | Gene names | Rims3, Kiaa0237 | |||
|
Domain Architecture |
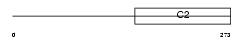
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.061856 (rank : 114) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.061856 (rank : 115) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.054549 (rank : 147) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.053148 (rank : 153) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.069319 (rank : 87) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.063570 (rank : 106) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
RSRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.081999 (rank : 55) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96IZ7, Q96QK2, Q9NZE5 | Gene names | RSRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
RSRC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.077319 (rank : 66) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DBU6, Q3TF06 | Gene names | Rsrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
RU17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.060446 (rank : 122) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
RU17_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.050266 (rank : 174) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.058733 (rank : 129) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
SCG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.070257 (rank : 85) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.182465 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.150561 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.154604 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SFR16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.070856 (rank : 82) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
SFR16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.072884 (rank : 78) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
SFR17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.127023 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.093645 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.096646 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.054355 (rank : 148) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
SFRS8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.053590 (rank : 150) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.059557 (rank : 127) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.054584 (rank : 146) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SNIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.052493 (rank : 161) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAD8, Q96SP9, Q9H9T7 | Gene names | SNIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
SNIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.052395 (rank : 163) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BIZ6, Q3V106, Q8BIZ4 | Gene names | Snip1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
SON_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.058268 (rank : 133) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SRCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.085000 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.088752 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.092889 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.068439 (rank : 90) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
T2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.099937 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
TAF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.056930 (rank : 138) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.061604 (rank : 119) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TARA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.070483 (rank : 83) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.053263 (rank : 152) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.052605 (rank : 157) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.063101 (rank : 108) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.094481 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TOP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.062385 (rank : 110) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
TOP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.060032 (rank : 124) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
TR150_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.050742 (rank : 171) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TR150_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.068149 (rank : 92) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.085599 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.071924 (rank : 81) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.051406 (rank : 166) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.081969 (rank : 56) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.055310 (rank : 143) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.056247 (rank : 140) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.059713 (rank : 126) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.059241 (rank : 128) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
WDR60_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.072172 (rank : 80) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
WDR60_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.074683 (rank : 75) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
XE7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.076644 (rank : 69) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.086786 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ZRAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.066710 (rank : 94) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
SNUT3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.01107e-24 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
SNUT3_MOUSE
|
||||||
| NC score | 0.969763 (rank : 2) | θ value | 4.01107e-24 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K194, Q8BRA8, Q9CSV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3. | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.248427 (rank : 3) | θ value | 0.00509761 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.239808 (rank : 4) | θ value | 0.00665767 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.213721 (rank : 5) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.182465 (rank : 6) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.179733 (rank : 7) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
RBM15_HUMAN
|
||||||
| NC score | 0.179035 (rank : 8) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.165707 (rank : 9) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.154604 (rank : 10) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.150561 (rank : 11) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.143756 (rank : 12) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.140592 (rank : 13) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.137294 (rank : 14) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.134045 (rank : 15) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.133288 (rank : 16) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.129834 (rank : 17) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.127731 (rank : 18) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.127458 (rank : 19) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.127023 (rank : 20) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.126121 (rank : 21) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.114867 (rank : 22) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.113983 (rank : 23) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.113201 (rank : 24) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.108336 (rank : 25) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.107107 (rank : 26) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.100464 (rank : 27) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
T2_MOUSE
|
||||||
| NC score | 0.099937 (rank : 28) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.099701 (rank : 29) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.099632 (rank : 30) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.098000 (rank : 31) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.096982 (rank : 32) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.096883 (rank : 33) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.096824 (rank : 34) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.096646 (rank : 35) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.096550 (rank : 36) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.096245 (rank : 37) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.096038 (rank : 38) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.094616 (rank : 39) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.094481 (rank : 40) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.093882 (rank : 41) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.093645 (rank : 42) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.092889 (rank : 43) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.089690 (rank : 44) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.088752 (rank : 45) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.086786 (rank : 46) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.085599 (rank : 47) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
LC7L2_MOUSE
|
||||||
| NC score | 0.085075 (rank : 48) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TNC4, Q99LM5, Q99PC3 | Gene names | Luc7l2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2 (CGI-74 homolog). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.085000 (rank : 49) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.084344 (rank : 50) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.083689 (rank : 51) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.083218 (rank : 52) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.083064 (rank : 53) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.082295 (rank : 54) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
RSRC1_HUMAN
|
||||||
| NC score | 0.081999 (rank : 55) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96IZ7, Q96QK2, Q9NZE5 | Gene names | RSRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.081969 (rank : 56) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.081674 (rank : 57) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
LC7L2_HUMAN
|
||||||
| NC score | 0.081109 (rank : 58) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y383, Q8IUP9, Q9NVL3, Q9NVN7, Q9UQN1 | Gene names | LUC7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 2. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.079807 (rank : 59) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.079246 (rank : 60) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
ADDG_MOUSE
|
||||||
| NC score | 0.078405 (rank : 61) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QYB5, Q9D2M5, Q9QYB6 | Gene names | Add3, Addl | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.078014 (rank : 62) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.077792 (rank : 63) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
ADDA_HUMAN
|
||||||
| NC score | 0.077611 (rank : 64) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35611, Q13734, Q14729, Q16156, Q9UJB6 | Gene names | ADD1, ADDA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.077559 (rank : 65) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
RSRC1_MOUSE
|
||||||
| NC score | 0.077319 (rank : 66) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DBU6, Q3TF06 | Gene names | Rsrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
ADDG_HUMAN
|
||||||
| NC score | 0.077307 (rank : 67) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UEY8, O43243, Q92773, Q9UEY7 | Gene names | ADD3, ADDL | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
REN3B_HUMAN
|
||||||
| NC score | 0.077197 (rank : 68) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BZI7, Q9H1J0 | Gene names | UPF3B, RENT3B, UPF3X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3B (Nonsense mRNA reducing factor 3B) (Up-frameshift suppressor 3 homolog B) (hUpf3B) (hUpf3p-X). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.076644 (rank : 69) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.076277 (rank : 70) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.075644 (rank : 71) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMN_HUMAN
|
||||||
| NC score | 0.075387 (rank : 72) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.075325 (rank : 73) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CA065_HUMAN
|
||||||
| NC score | 0.075111 (rank : 74) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
WDR60_MOUSE
|
||||||
| NC score | 0.074683 (rank : 75) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.074071 (rank : 76) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.073268 (rank : 77) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SFR16_MOUSE
|
||||||
| NC score | 0.072884 (rank : 78) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.072326 (rank : 79) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.072172 (rank : 80) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.071924 (rank : 81) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
SFR16_HUMAN
|
||||||
| NC score | 0.070856 (rank : 82) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N2M8, O96026, Q6UW71, Q96DX2 | Gene names | SFRS16, SWAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.070483 (rank : 83) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.070265 (rank : 84) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.070257 (rank : 85) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.070017 (rank : 86) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.069319 (rank : 87) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.068771 (rank : 88) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.068540 (rank : 89) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.068439 (rank : 90) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.068225 (rank : 91) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.068149 (rank : 92) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.067376 (rank : 93) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
ZRAB2_HUMAN
|
||||||
| NC score | 0.066710 (rank : 94) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.066656 (rank : 95) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.066333 (rank : 96) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.065943 (rank : 97) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.065466 (rank : 98) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.064895 (rank : 99) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.064883 (rank : 100) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.064593 (rank : 101) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.064235 (rank : 102) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CENPT_MOUSE
|
||||||
| NC score | 0.064039 (rank : 103) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.063849 (rank : 104) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.063761 (rank : 105) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.063570 (rank : 106) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.063287 (rank : 107) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.063101 (rank : 108) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.062881 (rank : 109) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
TOP1_HUMAN
|
||||||
| NC score | 0.062385 (rank : 110) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11387, O43256, Q12855, Q12856, Q15610, Q5TFY3, Q9UJN0 | Gene names | TOP1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.062289 (rank : 111) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.062035 (rank : 112) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.062027 (rank : 113) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.061856 (rank : 114) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.061856 (rank : 115) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.061724 (rank : 116) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.061708 (rank : 117) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.061651 (rank : 118) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.061604 (rank : 119) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.061033 (rank : 120) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.060908 (rank : 121) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
RU17_HUMAN
|
||||||
| NC score | 0.060446 (rank : 122) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08621, P78493, P78494, Q15364, Q15686, Q15687, Q15689, Q99377, Q9UE45, Q9UE46, Q9UE47, Q9UE48, Q9UFQ6 | Gene names | SNRP70, RPU1, U1AP1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 snRNP 70 kDa) (snRNP70) (U1-70K). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.060367 (rank : 123) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
TOP1_MOUSE
|
||||||
| NC score | 0.060032 (rank : 124) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04750 | Gene names | Top1, Top-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 1 (EC 5.99.1.2) (DNA topoisomerase I). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.059743 (rank : 125) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.059713 (rank : 126) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.059557 (rank : 127) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.059241 (rank : 128) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.058733 (rank : 129) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.058708 (rank : 130) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.058596 (rank : 131) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.058296 (rank : 132) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.058268 (rank : 133) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.057738 (rank : 134) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CS029_MOUSE
|
||||||
| NC score | 0.057734 (rank : 135) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.057708 (rank : 136) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.057320 (rank : 137) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.056930 (rank : 138) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
FIP1_MOUSE
|
||||||
| NC score | 0.056590 (rank : 139) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D824, Q8BWX7, Q99LH0, Q9DBB2 | Gene names | Fip1l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA 3'-end-processing factor FIP1 (FIP1-like 1). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.056247 (rank : 140) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.055865 (rank : 141) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.055626 (rank : 142) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.055310 (rank : 143) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.055245 (rank : 144) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.055206 (rank : 145) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.054584 (rank : 146) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.054549 (rank : 147) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.054355 (rank : 148) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.054172 (rank : 149) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
SFRS8_HUMAN
|
||||||
| NC score | 0.053590 (rank : 150) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12872 | Gene names | SFRS8, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 8 (Suppressor of white apricot protein homolog). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.053504 (rank : 151) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.053263 (rank : 152) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.053148 (rank : 153) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.053062 (rank : 154) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.052867 (rank : 155) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.052641 (rank : 156) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.052605 (rank : 157) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.052577 (rank : 158) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
RIMS3_HUMAN
|
||||||
| NC score | 0.052504 (rank : 159) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.052500 (rank : 160) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
SNIP1_HUMAN
|
||||||
| NC score | 0.052493 (rank : 161) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAD8, Q96SP9, Q9H9T7 | Gene names | SNIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
RIMS3_MOUSE
|
||||||
| NC score | 0.052457 (rank : 162) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U57 | Gene names | Rims3, Kiaa0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
SNIP1_MOUSE
|
||||||
| NC score | 0.052395 (rank : 163) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BIZ6, Q3V106, Q8BIZ4 | Gene names | Snip1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.052135 (rank : 164) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.051898 (rank : 165) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.051406 (rank : 166) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.051289 (rank : 167) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.051184 (rank : 168) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.051097 (rank : 169) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.050747 (rank : 170) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TR150_HUMAN
|
||||||
| NC score | 0.050742 (rank : 171) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2W1, Q5VTK6 | Gene names | THRAP3, TRAP150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.050487 (rank : 172) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.050453 (rank : 173) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RU17_MOUSE
|
||||||
| NC score | 0.050266 (rank : 174) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62376, Q3UIW4 | Gene names | Snrp70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U1 small nuclear ribonucleoprotein 70 kDa (U1 SNRNP 70 kDa) (snRNP70). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.049530 (rank : 175) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LRC27_HUMAN
|
||||||
| NC score | 0.047751 (rank : 176) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0I9, Q5SZH6, Q5SZH8, Q5SZH9, Q86XT5, Q8N7C8, Q8NA21 | Gene names | LRRC27, KIAA1674 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.047066 (rank : 177) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
DCDC2_MOUSE
|
||||||
| NC score | 0.042980 (rank : 178) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DU00, Q5SZU0, Q80Y99 | Gene names | Dcdc2, Kiaa1154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublecortin domain-containing protein 2. | |||||
|
DLG5_HUMAN
|
||||||
| NC score | 0.041350 (rank : 179) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.041192 (rank : 180) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.038794 (rank : 181) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.037361 (rank : 182) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.035484 (rank : 183) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
TBX2_HUMAN
|
||||||
| NC score | 0.035088 (rank : 184) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13207, Q16424, Q7Z647 | Gene names | TBX2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.027248 (rank : 185) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.022425 (rank : 186) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SEPT3_HUMAN
|
||||||
| NC score | 0.020140 (rank : 187) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UH03, Q6IBZ6, Q8N3P3 | Gene names | SEPT3, SEP3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal-specific septin-3. | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.019771 (rank : 188) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.014205 (rank : 189) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | 0.010848 (rank : 190) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||