Please be patient as the page loads
|
CENPT_MOUSE
|
||||||
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CENPT_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
CENPT_HUMAN
|
||||||
| θ value | 8.67031e-128 (rank : 2) | NC score | 0.913983 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96BT3, Q96I29, Q96IC6, Q96NK9, Q9H901 | Gene names | CENPT, C16orf56, ICEN22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T) (Interphase centromere complex protein 22). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 3) | NC score | 0.094167 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 4) | NC score | 0.065516 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 5) | NC score | 0.084113 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.062215 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.055012 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.126300 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TSH3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.050588 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.119889 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.089323 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.127158 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.039620 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.117524 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.074728 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.070304 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ZO1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.035596 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.078770 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
B3A3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.027622 (rank : 135) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.099814 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CENPS_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.156138 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N2Z9, Q8NFE5, Q8NFG5 | Gene names | APITD1, CENPS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein S (CENP-S) (Apoptosis-inducing TAF9-like domain- containing protein 1). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.068742 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
CENPS_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.148456 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D084, Q80ZS2, Q8BH28 | Gene names | Apitd1, Cenps | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein S (CENP-S) (Apoptosis-inducing TAF9-like domain- containing protein 1 homolog). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.028177 (rank : 134) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
KLF17_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.011203 (rank : 170) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.046044 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.085876 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.050344 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.071727 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.045166 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
ZDHC8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.030417 (rank : 129) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.046534 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.064538 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.042269 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.018306 (rank : 153) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.065659 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.049320 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.055433 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.068346 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.046686 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.040168 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.107964 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.063674 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.064761 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.054063 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SNUT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.064039 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.017722 (rank : 154) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
BAT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.062078 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.051396 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CN021_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.031661 (rank : 126) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86U38, Q8IVF0, Q8TBS6 | Gene names | C14orf21, KIAA2021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf21. | |||||
|
GCYA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.015204 (rank : 162) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
HXC10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.016139 (rank : 157) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31257, P31312, Q7TMT7 | Gene names | Hoxc10, Hox-3.6, Hoxc-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-C10 (Hox-3.6). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.050736 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.024484 (rank : 140) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.021250 (rank : 145) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NID1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.009454 (rank : 173) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
PHX2A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.011977 (rank : 169) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14813, Q8IVZ2 | Gene names | PHOX2A, ARIX, PMX2A | |||
|
Domain Architecture |
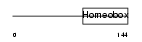
|
|||||
| Description | Paired mesoderm homeobox protein 2A (Paired-like homeobox 2A) (Aristaless homeobox protein homolog) (ARIX1 homeodomain protein). | |||||
|
SFRS2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.029615 (rank : 131) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |
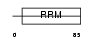
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.029615 (rank : 132) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |
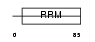
|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
SIGL5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.016222 (rank : 156) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15389 | Gene names | SIGLEC5, CD33L2, OBBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 5 precursor (Siglec-5) (Obesity- binding protein 2) (OB-binding protein 2) (OB-BP2) (CD33 antigen-like 2) (CD170 antigen). | |||||
|
SON_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.069572 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SRR35_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.025595 (rank : 138) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |
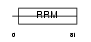
|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
STC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.029753 (rank : 130) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76061 | Gene names | STC2 | |||
|
Domain Architecture |

|
|||||
| Description | Stanniocalcin-2 precursor (STC-2) (Stanniocalcin-related protein) (STCRP) (STC-related protein). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.011032 (rank : 171) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.015770 (rank : 160) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
NRBP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.019217 (rank : 152) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHY1, Q53FZ5, Q96SU3 | Gene names | NRBP1, BCON3, NRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding protein. | |||||
|
NRBP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.019393 (rank : 150) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99J45, Q8BL77 | Gene names | Nrbp1, Madm, Nrbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding protein (MLF1 adapter molecule) (HLS7- interacting protein kinase). | |||||
|
PAX3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.013424 (rank : 166) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-3. | |||||
|
PTN18_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.009436 (rank : 174) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.029496 (rank : 133) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
T3JAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.016434 (rank : 155) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
TNR21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.020452 (rank : 147) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
ABI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.013098 (rank : 167) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P62484, Q6PHU3 | Gene names | Abi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.020237 (rank : 148) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
CCD96_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.031514 (rank : 127) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CD248_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.023933 (rank : 143) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CENPA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.052029 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49450, Q53T74, Q9BVW2 | Gene names | CENPA | |||
|
Domain Architecture |
|
|||||
| Description | Histone H3-like centromeric protein A (Centromere protein A) (CENP-A) (Centromere autoantigen A). | |||||
|
CP007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.027221 (rank : 136) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.054527 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FOXK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.014327 (rank : 163) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
FOXO3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.013852 (rank : 164) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43524, O15171, Q5T2I7, Q9BZ04 | Gene names | FOXO3A, FKHRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O3A (Forkhead in rhabdomyosarcoma-like 1) (AF6q21 protein). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.056071 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PANK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.015540 (rank : 161) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.075601 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.009630 (rank : 172) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.031036 (rank : 128) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.004310 (rank : 178) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
CBL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.023963 (rank : 142) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
CS029_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.024977 (rank : 139) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
CSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.033188 (rank : 125) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
|
DACH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.019376 (rank : 151) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.027100 (rank : 137) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.008242 (rank : 175) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.033727 (rank : 123) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
LUC7L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.016112 (rank : 158) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ29, Q96S32, Q9NPH4 | Gene names | LUC7L, LUC7L1 | |||
|
Domain Architecture |
|
|||||
| Description | Putative RNA-binding protein Luc7-like 1 (SR+89) (Putative SR protein LUC7B1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.053097 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.006166 (rank : 177) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
PAX3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.012663 (rank : 168) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.007069 (rank : 176) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.045921 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.075122 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.035775 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.037152 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.050842 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.055991 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.047291 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.021013 (rank : 146) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HXC10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.013506 (rank : 165) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
MKL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.023689 (rank : 144) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
SMG5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.016056 (rank : 159) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
SUPT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.024078 (rank : 141) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75486, O76066 | Gene names | SUPT3H, SPT3 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription initiation protein SPT3 homolog (SPT3-like protein). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.003710 (rank : 179) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
WASF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.033669 (rank : 124) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.034090 (rank : 122) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.020065 (rank : 149) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ALEX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.060578 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.054003 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.086466 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051665 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052874 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.055092 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.071414 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.052042 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.073184 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.061066 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.060705 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.051933 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.055034 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.068109 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.061229 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.057609 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.055949 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.074224 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.066976 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.067115 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.063359 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.054443 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.059009 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.054967 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.087145 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.058189 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054002 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.052300 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.051852 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.052283 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PO121_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.054571 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.065452 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.064710 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.065277 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.053780 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.077443 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.082990 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.055610 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.063092 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.051145 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RERE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.050794 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.053519 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SELPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.050969 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
SELV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.058357 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.059793 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.053649 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.068660 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.082629 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.050162 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.069042 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TARA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.060266 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.050927 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054097 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.056666 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.065320 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.053175 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.064577 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.064099 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.057262 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.066644 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.064489 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.054128 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.055039 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052872 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CENPT_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
CENPT_HUMAN
|
||||||
| NC score | 0.913983 (rank : 2) | θ value | 8.67031e-128 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96BT3, Q96I29, Q96IC6, Q96NK9, Q9H901 | Gene names | CENPT, C16orf56, ICEN22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T) (Interphase centromere complex protein 22). | |||||
|
CENPS_HUMAN
|
||||||
| NC score | 0.156138 (rank : 3) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N2Z9, Q8NFE5, Q8NFG5 | Gene names | APITD1, CENPS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein S (CENP-S) (Apoptosis-inducing TAF9-like domain- containing protein 1). | |||||
|
CENPS_MOUSE
|
||||||
| NC score | 0.148456 (rank : 4) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D084, Q80ZS2, Q8BH28 | Gene names | Apitd1, Cenps | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein S (CENP-S) (Apoptosis-inducing TAF9-like domain- containing protein 1 homolog). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.127158 (rank : 5) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.126300 (rank : 6) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.119889 (rank : 7) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.117524 (rank : 8) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.107964 (rank : 9) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.099814 (rank : 10) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.094167 (rank : 11) | θ value | 0.00298849 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.089323 (rank : 12) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.087145 (rank : 13) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.086466 (rank : 14) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.085876 (rank : 15) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.084113 (rank : 16) | θ value | 0.0252991 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.082990 (rank : 17) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.082629 (rank : 18) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.078770 (rank : 19) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.077443 (rank : 20) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.075601 (rank : 21) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.075122 (rank : 22) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.074728 (rank : 23) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.074224 (rank : 24) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.073184 (rank : 25) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.071727 (rank : 26) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.071414 (rank : 27) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.070304 (rank : 28) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.069572 (rank : 29) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.069042 (rank : 30) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.068742 (rank : 31) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.068660 (rank : 32) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.068346 (rank : 33) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.068109 (rank : 34) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.067115 (rank : 35) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.066976 (rank : 36) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.066644 (rank : 37) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.065659 (rank : 38) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.065516 (rank : 39) | θ value | 0.00390308 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.065452 (rank : 40) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.065320 (rank : 41) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.065277 (rank : 42) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.064761 (rank : 43) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.064710 (rank : 44) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.064577 (rank : 45) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.064538 (rank : 46) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.064489 (rank : 47) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.064099 (rank : 48) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
SNUT3_HUMAN
|
||||||
| NC score | 0.064039 (rank : 49) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WVK2, Q15410 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 3 (U4/U6.U5 tri-snRNP-associated 27 kDa protein) (27K) (Nucleic acid-binding protein RY-1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.063674 (rank : 50) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.063359 (rank : 51) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.063092 (rank : 52) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.062215 (rank : 53) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BAT3_HUMAN
|
||||||
| NC score | 0.062078 (rank : 54) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P46379, O95874, Q5SQ35, Q5SQ36, Q5SQ37, Q5SRP8, Q96SA6, Q9BCN4 | Gene names | BAT3, G3 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3) (Protein G3). | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.061229 (rank : 55) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.061066 (rank : 56) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.060705 (rank : 57) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.060578 (rank : 58) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.060266 (rank : 59) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.059793 (rank : 60) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.059009 (rank : 61) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.058357 (rank : 62) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.058189 (rank : 63) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.057609 (rank : 64) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.057262 (rank : 65) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.056666 (rank : 66) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.056071 (rank : 67) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.055991 (rank : 68) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.055949 (rank : 69) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.055610 (rank : 70) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.055433 (rank : 71) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.055092 (rank : 72) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.055039 (rank : 73) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.055034 (rank : 74) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.055012 (rank : 75) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.054967 (rank : 76) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.054571 (rank : 77) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.054527 (rank : 78) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.054443 (rank : 79) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.054128 (rank : 80) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.054097 (rank : 81) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.054063 (rank : 82) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.054003 (rank : 83) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.054002 (rank : 84) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.053780 (rank : 85) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.053649 (rank : 86) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.053519 (rank : 87) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.053175 (rank : 88) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.053097 (rank : 89) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.052874 (rank : 90) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.052872 (rank : 91) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.052300 (rank : 92) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.052283 (rank : 93) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.052042 (rank : 94) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
CENPA_HUMAN
|
||||||
| NC score | 0.052029 (rank : 95) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49450, Q53T74, Q9BVW2 | Gene names | CENPA | |||
|
Domain Architecture |

|
|||||
| Description | Histone H3-like centromeric protein A (Centromere protein A) (CENP-A) (Centromere autoantigen A). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.051933 (rank : 96) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.051852 (rank : 97) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.051665 (rank : 98) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.051396 (rank : 99) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.051145 (rank : 100) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.050969 (rank : 101) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.050927 (rank : 102) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.050842 (rank : 103) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.050794 (rank : 104) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.050736 (rank : 105) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
TSH3_MOUSE
|
||||||
| NC score | 0.050588 (rank : 106) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.050344 (rank : 107) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.050162 (rank : 108) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.049320 (rank : 109) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.047291 (rank : 110) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.046686 (rank : 111) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.046534 (rank : 112) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.046044 (rank : 113) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.045921 (rank : 114) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.045166 (rank : 115) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.042269 (rank : 116) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.040168 (rank : 117) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.039620 (rank : 118) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.037152 (rank : 119) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.035775 (rank : 120) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.035596 (rank : 121) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.034090 (rank : 122) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.033727 (rank : 123) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
WASF3_MOUSE
|
||||||
| NC score | 0.033669 (rank : 124) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
CSF1_MOUSE
|
||||||
| NC score | 0.033188 (rank : 125) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07141, Q8R3C8 | Gene names | Csf1, Csfm | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF). | |||||
|
CN021_HUMAN
|
||||||
| NC score | 0.031661 (rank : 126) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86U38, Q8IVF0, Q8TBS6 | Gene names | C14orf21, KIAA2021 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf21. | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.031514 (rank : 127) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.031036 (rank : 128) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ZDHC8_MOUSE
|
||||||
| NC score | 0.030417 (rank : 129) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
STC2_HUMAN
|
||||||
| NC score | 0.029753 (rank : 130) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76061 | Gene names | STC2 | |||
|
Domain Architecture |

|
|||||
| Description | Stanniocalcin-2 precursor (STC-2) (Stanniocalcin-related protein) (STCRP) (STC-related protein). | |||||
|
SFRS2_HUMAN
|
||||||
| NC score | 0.029615 (rank : 131) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01130 | Gene names | SFRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264). | |||||
|
SFRS2_MOUSE
|
||||||
| NC score | 0.029615 (rank : 132) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62093, Q542L3, Q60701 | Gene names | Sfrs2, Pr264, Sfrs10 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 2 (Splicing factor SC35) (SC-35) (Splicing component, 35 kDa) (Protein PR264) (Putative myelin regulatory factor 1) (MRF-1). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.029496 (rank : 133) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.028177 (rank : 134) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
B3A3_MOUSE
|
||||||
| NC score | 0.027622 (rank : 135) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
CP007_MOUSE
|
||||||
| NC score | 0.027221 (rank : 136) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.027100 (rank : 137) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
SRR35_HUMAN
|
||||||
| NC score | 0.025595 (rank : 138) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXF0, Q8WW25 | Gene names | SRRP35 | |||
|
Domain Architecture |

|
|||||
| Description | 35 kDa SR repressor protein (SRrp35). | |||||
|
CS029_MOUSE
|
||||||
| NC score | 0.024977 (rank : 139) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.024484 (rank : 140) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
SUPT3_HUMAN
|
||||||
| NC score | 0.024078 (rank : 141) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75486, O76066 | Gene names | SUPT3H, SPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation protein SPT3 homolog (SPT3-like protein). | |||||
|
CBL_MOUSE
|
||||||
| NC score | 0.023963 (rank : 142) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.023933 (rank : 143) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
MKL2_HUMAN
|
||||||
| NC score | 0.023689 (rank : 144) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.021250 (rank : 145) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.021013 (rank : 146) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
TNR21_MOUSE
|
||||||
| NC score | 0.020452 (rank : 147) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.020237 (rank : 148) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.020065 (rank : 149) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
NRBP_MOUSE
|
||||||
| NC score | 0.019393 (rank : 150) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99J45, Q8BL77 | Gene names | Nrbp1, Madm, Nrbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding protein (MLF1 adapter molecule) (HLS7- interacting protein kinase). | |||||
|
DACH1_HUMAN
|
||||||
| NC score | 0.019376 (rank : 151) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
NRBP_HUMAN
|
||||||
| NC score | 0.019217 (rank : 152) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHY1, Q53FZ5, Q96SU3 | Gene names | NRBP1, BCON3, NRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding protein. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.018306 (rank : 153) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.017722 (rank : 154) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
T3JAM_MOUSE
|
||||||
| NC score | 0.016434 (rank : 155) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0G2, Q8BY50, Q8K1Y9 | Gene names | Traf3ip3, T3jam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRAF3-interacting JNK-activating modulator (TRAF3-interacting protein 3). | |||||
|
SIGL5_HUMAN
|
||||||
| NC score | 0.016222 (rank : 156) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15389 | Gene names | SIGLEC5, CD33L2, OBBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 5 precursor (Siglec-5) (Obesity- binding protein 2) (OB-binding protein 2) (OB-BP2) (CD33 antigen-like 2) (CD170 antigen). | |||||
|
HXC10_MOUSE
|
||||||
| NC score | 0.016139 (rank : 157) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31257, P31312, Q7TMT7 | Gene names | Hoxc10, Hox-3.6, Hoxc-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein Hox-C10 (Hox-3.6). | |||||
|
LUC7L_HUMAN
|
||||||
| NC score | 0.016112 (rank : 158) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ29, Q96S32, Q9NPH4 | Gene names | LUC7L, LUC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein Luc7-like 1 (SR+89) (Putative SR protein LUC7B1). | |||||
|
SMG5_MOUSE
|
||||||
| NC score | 0.016056 (rank : 159) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZPY2, Q497S2, Q8R3S4 | Gene names | Smg5, Est1b, Kiaa1089 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SMG5 (SMG-5 homolog) (EST1-like protein B). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.015770 (rank : 160) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
PANK1_MOUSE
|
||||||
| NC score | 0.015540 (rank : 161) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
GCYA2_HUMAN
|
||||||
| NC score | 0.015204 (rank : 162) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P33402 | Gene names | GUCY1A2, GUC1A2, GUCSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate cyclase soluble subunit alpha-2 (EC 4.6.1.2) (GCS-alpha-2). | |||||
|
FOXK2_HUMAN
|
||||||
| NC score | 0.014327 (rank : 163) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
FOXO3_HUMAN
|
||||||
| NC score | 0.013852 (rank : 164) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43524, O15171, Q5T2I7, Q9BZ04 | Gene names | FOXO3A, FKHRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O3A (Forkhead in rhabdomyosarcoma-like 1) (AF6q21 protein). | |||||
|
HXC10_HUMAN
|
||||||
| NC score | 0.013506 (rank : 165) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
PAX3_MOUSE
|
||||||
| NC score | 0.013424 (rank : 166) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3. | |||||
|
ABI2_MOUSE
|
||||||
| NC score | 0.013098 (rank : 167) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P62484, Q6PHU3 | Gene names | Abi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2). | |||||
|
PAX3_HUMAN
|
||||||
| NC score | 0.012663 (rank : 168) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
PHX2A_HUMAN
|
||||||
| NC score | 0.011977 (rank : 169) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14813, Q8IVZ2 | Gene names | PHOX2A, ARIX, PMX2A | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2A (Paired-like homeobox 2A) (Aristaless homeobox protein homolog) (ARIX1 homeodomain protein). | |||||
|
KLF17_HUMAN
|
||||||
| NC score | 0.011203 (rank : 170) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.011032 (rank : 171) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.009630 (rank : 172) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
NID1_HUMAN
|
||||||
| NC score | 0.009454 (rank : 173) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.009436 (rank : 174) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.008242 (rank : 175) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.007069 (rank : 176) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.006166 (rank : 177) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.004310 (rank : 178) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.003710 (rank : 179) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||