Please be patient as the page loads
|
SAPS1_MOUSE
|
||||||
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.848013 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
SAPS3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.960233 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
SAPS3_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.958997 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
SAPS2_HUMAN
|
||||||
| θ value | 1.55827e-177 (rank : 5) | NC score | 0.959227 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
SAPS2_MOUSE
|
||||||
| θ value | 1.31915e-176 (rank : 6) | NC score | 0.955748 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3Q2, Q9CXB7 | Gene names | Saps2, Kiaa0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 7) | NC score | 0.081979 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 8) | NC score | 0.078422 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.014417 (rank : 146) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.086528 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.050704 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.047282 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.087845 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.046227 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.041241 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.022537 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.092694 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.090648 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.032231 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.037072 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.127621 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.063185 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.095993 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.080537 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.039873 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
MAGAA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.023667 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.063611 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.047496 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.034609 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
HEAT1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.046317 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H583, Q5T3Q8, Q9NW23 | Gene names | HEATR1, BAP28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 1 (Protein BAP28). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.034068 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.041007 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.061853 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
FRS3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.029946 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.053624 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
TM108_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.047592 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
NBN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.027375 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.021619 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PCD12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.004583 (rank : 176) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.067788 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.033531 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.038443 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.027676 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.093913 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.009460 (rank : 159) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TLE2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.014637 (rank : 145) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVB2 | Gene names | Tle2, Grg2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2. | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.062676 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.038763 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
C1QR1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.009854 (rank : 158) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.024615 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.040699 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.034532 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.004833 (rank : 174) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.040691 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.026995 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.029703 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
GT2D1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.018533 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHL9, O95444, Q75MX7, Q86UM3, Q8WVC4, Q9UHK8, Q9UI91 | Gene names | GTF2IRD1, CREAM1, GTF3, MUSTRD1, RBAP2, WBSCR11, WBSCR12 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Muscle TFII-I repeat domain-containing protein 1) (General transcription factor III) (Slow- muscle-fiber enhancer-binding protein) (USE B1-binding protein) (MusTRD1/BEN) (Williams-Beuren syndrome chromosome region 11 protein). | |||||
|
PAX6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.010406 (rank : 155) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26367, Q6N006, Q99413 | Gene names | PAX6, AN2 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-6 (Oculorhombin) (Aniridia type II protein). | |||||
|
PAX6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.010406 (rank : 156) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P63015, P32117, P70601, Q62222, Q64037, Q8CEI5, Q8VDB5, Q921Q8 | Gene names | Pax6, Pax-6, Sey | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-6 (Oculorhombin). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.073378 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.066483 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.032381 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
BCL6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.001457 (rank : 182) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
FRS3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.028486 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.004668 (rank : 175) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYL6B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.008169 (rank : 164) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.055469 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PCGF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.024854 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
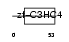
|
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.063203 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
TLE2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.015288 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.059928 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.025847 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
K1688_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.021425 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.012533 (rank : 149) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.048441 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PCGF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.022901 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
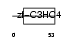
|
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
PKHG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.011045 (rank : 153) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.059277 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.023844 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
CENPT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.029496 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.016257 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.041031 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
IC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.011966 (rank : 152) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.006698 (rank : 167) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LARP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.025870 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.059509 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MY116_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.027565 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17564 | Gene names | Myd116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid differentiation primary response protein MyD116. | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.047229 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
POLH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.013370 (rank : 147) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJN0, Q9JJJ2 | Gene names | Polh, Rad30a, Xpv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein homolog). | |||||
|
STON2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.016975 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.010925 (rank : 154) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
APC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.034124 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
ARHGA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.009097 (rank : 160) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.062957 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DLGP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.018505 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.007445 (rank : 166) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
FOXD3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.003972 (rank : 177) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJU5, Q9BYM2, Q9UDD1 | Gene names | FOXD3 | |||
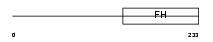
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.026913 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.021881 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.027274 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.027313 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.030561 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
SOX7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.006023 (rank : 169) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.003321 (rank : 180) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.046801 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
ANKY2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.006666 (rank : 168) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.035137 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CN004_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.020048 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
CN004_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.022749 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.035774 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
EGF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.003471 (rank : 179) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
FA13C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.012126 (rank : 151) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
FOXN4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.005221 (rank : 171) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Q3, Q920C0 | Gene names | Foxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N4. | |||||
|
GBX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.004923 (rank : 173) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52951, O43833, Q53RX5, Q9Y5Y1 | Gene names | GBX2 | |||
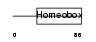
|
Domain Architecture |
|
|||||
| Description | Homeobox protein GBX-2 (Gastrulation and brain-specific homeobox protein 2). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.026334 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
ICAL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.021230 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.060792 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NKX31_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.005110 (rank : 172) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97436, O09087 | Gene names | Nkx3-1, Nkx-3.1, Nkx3a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.1. | |||||
|
NPHP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.012397 (rank : 150) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75161 | Gene names | NPHP4, KIAA0673 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
RANB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.020402 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
SN1L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | -0.000433 (rank : 184) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60670 | Gene names | Snf1lk, Msk | |||
|
Domain Architecture |
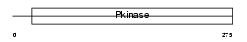
|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK) (HRT-20) (Myocardial SNF1- like kinase). | |||||
|
SON_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.045929 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SYN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.030303 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.041143 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.012774 (rank : 148) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
VGFR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | -0.000368 (rank : 183) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35917 | Gene names | Flt4, Flt-4 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 3 precursor (EC 2.7.10.1) (VEGFR-3) (Tyrosine-protein kinase receptor FLT4). | |||||
|
ZFP95_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | -0.000859 (rank : 186) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |
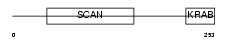
|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||
|
ZSWM4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.008197 (rank : 163) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C7B8, Q8BPN9 | Gene names | Zswim4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 4. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.002227 (rank : 181) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
HIRA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.003907 (rank : 178) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |
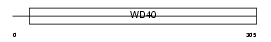
|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.023938 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.022547 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.050578 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
KPB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.016536 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46020, Q2M3D7 | Gene names | PHKA1, PHKA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform (Phosphorylase kinase alpha M subunit). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.022722 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.074875 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.008411 (rank : 161) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.014751 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
MYPN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.008354 (rank : 162) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.007964 (rank : 165) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
SNRK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | -0.000704 (rank : 185) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRH2, Q14706, Q68D15, Q6IQ46, Q9NXI7 | Gene names | SNRK, KIAA0096, SNFRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
TRI37_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.005362 (rank : 170) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PCX9, Q8CHC5 | Gene names | Trim37, Kiaa0898 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37. | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.018106 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.010263 (rank : 157) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.060325 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.050701 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.050959 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.055703 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.059833 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.053570 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.052308 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.054618 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.058259 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EP300_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.078377 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.050467 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.057316 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.051552 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.064894 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.081451 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.053801 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.054595 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.054166 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.059885 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.060814 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.081061 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.071968 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.093531 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.094135 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.050243 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.064789 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RERE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.057701 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SELPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.057734 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
SF01_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.050799 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.067856 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.060876 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.061231 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.063885 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.064288 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SYN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.062270 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.054918 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TM108_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.066827 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.071881 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.065408 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.074061 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.066211 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.051402 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
SAPS3_HUMAN
|
||||||
| NC score | 0.960233 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
SAPS2_HUMAN
|
||||||
| NC score | 0.959227 (rank : 3) | θ value | 1.55827e-177 (rank : 5) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
SAPS3_MOUSE
|
||||||
| NC score | 0.958997 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
SAPS2_MOUSE
|
||||||
| NC score | 0.955748 (rank : 5) | θ value | 1.31915e-176 (rank : 6) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3Q2, Q9CXB7 | Gene names | Saps2, Kiaa0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.848013 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.127621 (rank : 7) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.095993 (rank : 8) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.094135 (rank : 9) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.093913 (rank : 10) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.093531 (rank : 11) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.092694 (rank : 12) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.090648 (rank : 13) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.087845 (rank : 14) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.086528 (rank : 15) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.081979 (rank : 16) | θ value | 0.000461057 (rank : 7) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.081451 (rank : 17) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.081061 (rank : 18) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.080537 (rank : 19) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.078422 (rank : 20) | θ value | 0.00102713 (rank : 8) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.078377 (rank : 21) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.074875 (rank : 22) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.074061 (rank : 23) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.073378 (rank : 24) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.071968 (rank : 25) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.071881 (rank : 26) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.067856 (rank : 27) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.067788 (rank : 28) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.066827 (rank : 29) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.066483 (rank : 30) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.066211 (rank : 31) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.065408 (rank : 32) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.064894 (rank : 33) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.064789 (rank : 34) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.064288 (rank : 35) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.063885 (rank : 36) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.063611 (rank : 37) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.063203 (rank : 38) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.063185 (rank : 39) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.062957 (rank : 40) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.062676 (rank : 41) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.062270 (rank : 42) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.061853 (rank : 43) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.061231 (rank : 44) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.060876 (rank : 45) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.060814 (rank : 46) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.060792 (rank : 47) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.060325 (rank : 48) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.059928 (rank : 49) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.059885 (rank : 50) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.059833 (rank : 51) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.059509 (rank : 52) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.059277 (rank : 53) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.058259 (rank : 54) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.057734 (rank : 55) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.057701 (rank : 56) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.057316 (rank : 57) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.055703 (rank : 58) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.055469 (rank : 59) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.054918 (rank : 60) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.054618 (rank : 61) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.054595 (rank : 62) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.054166 (rank : 63) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.053801 (rank : 64) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.053624 (rank : 65) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.053570 (rank : 66) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.052308 (rank : 67) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.051552 (rank : 68) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.051402 (rank : 69) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.050959 (rank : 70) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.050799 (rank : 71) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.050704 (rank : 72) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.050701 (rank : 73) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.050578 (rank : 74) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.050467 (rank : 75) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.050243 (rank : 76) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.048441 (rank : 77) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TM108_HUMAN
|
||||||
| NC score | 0.047592 (rank : 78) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.047496 (rank : 79) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.047282 (rank : 80) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.047229 (rank : 81) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.046801 (rank : 82) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
HEAT1_HUMAN
|
||||||
| NC score | 0.046317 (rank : 83) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H583, Q5T3Q8, Q9NW23 | Gene names | HEATR1, BAP28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 1 (Protein BAP28). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.046227 (rank : 84) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.045929 (rank : 85) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.041241 (rank : 86) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.041143 (rank : 87) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.041031 (rank : 88) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.041007 (rank : 89) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.040699 (rank : 90) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.040691 (rank : 91) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.039873 (rank : 92) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.038763 (rank : 93) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.038443 (rank : 94) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.037072 (rank : 95) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.035774 (rank : 96) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.035137 (rank : 97) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.034609 (rank : 98) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.034532 (rank : 99) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.034124 (rank : 100) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.034068 (rank : 101) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.033531 (rank : 102) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.032381 (rank : 103) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.032231 (rank : 104) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.030561 (rank : 105) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
SYN2_MOUSE
|
||||||
| NC score | 0.030303 (rank : 106) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
FRS3_HUMAN
|
||||||
| NC score | 0.029946 (rank : 107) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43559, Q5T3D5 | Gene names | FRS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (Suc1-associated neurotrophic factor target 2) (SNT-2) (FGFR signaling adaptor SNT2). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.029703 (rank : 108) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
CENPT_MOUSE
|
||||||
| NC score | 0.029496 (rank : 109) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TJM4, Q3TPY6, Q3UPD2, Q8BTH0, Q8BTP2, Q8R5E9 | Gene names | Cenpt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein T (CENP-T). | |||||
|
FRS3_MOUSE
|
||||||
| NC score | 0.028486 (rank : 110) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.027676 (rank : 111) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
MY116_MOUSE
|
||||||
| NC score | 0.027565 (rank : 112) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P17564 | Gene names | Myd116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloid differentiation primary response protein MyD116. | |||||
|
NBN_HUMAN
|
||||||
| NC score | 0.027375 (rank : 113) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60934, O60672, Q32NF7, Q53FM6, Q63HR6, Q7LDM2 | Gene names | NBN, NBS, NBS1, P95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1) (Cell cycle regulatory protein p95). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.027313 (rank : 114) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.027274 (rank : 115) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.026995 (rank : 116) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.026913 (rank : 117) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.026334 (rank : 118) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LARP4_MOUSE
|
||||||
| NC score | 0.025870 (rank : 119) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.025847 (rank : 120) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
PCGF2_HUMAN
|
||||||
| NC score | 0.024854 (rank : 121) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.024615 (rank : 122) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.023938 (rank : 123) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.023844 (rank : 124) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
MAGAA_HUMAN
|
||||||
| NC score | 0.023667 (rank : 125) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P43363 | Gene names | MAGEA10, MAGE10 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 10 (MAGE-10 antigen). | |||||
|
PCGF2_MOUSE
|
||||||
| NC score | 0.022901 (rank : 126) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.022749 (rank : 127) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.022722 (rank : 128) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.022547 (rank : 129) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.022537 (rank : 130) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.021881 (rank : 131) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.021619 (rank : 132) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
K1688_MOUSE
|
||||||
| NC score | 0.021425 (rank : 133) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P59281, Q69ZD4, Q6P9R6 | Gene names | Kiaa1688, D15Wsu169e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1688. | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.021230 (rank : 134) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
RANB3_MOUSE
|
||||||
| NC score | 0.020402 (rank : 135) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.020048 (rank : 136) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
GT2D1_HUMAN
|
||||||
| NC score | 0.018533 (rank : 137) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHL9, O95444, Q75MX7, Q86UM3, Q8WVC4, Q9UHK8, Q9UI91 | Gene names | GTF2IRD1, CREAM1, GTF3, MUSTRD1, RBAP2, WBSCR11, WBSCR12 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Muscle TFII-I repeat domain-containing protein 1) (General transcription factor III) (Slow- muscle-fiber enhancer-binding protein) (USE B1-binding protein) (MusTRD1/BEN) (Williams-Beuren syndrome chromosome region 11 protein). | |||||
|
DLGP4_HUMAN
|
||||||
| NC score | 0.018505 (rank : 138) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.018106 (rank : 139) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
STON2_HUMAN
|
||||||
| NC score | 0.016975 (rank : 140) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
KPB1_HUMAN
|
||||||
| NC score | 0.016536 (rank : 141) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46020, Q2M3D7 | Gene names | PHKA1, PHKA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform (Phosphorylase kinase alpha M subunit). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.016257 (rank : 142) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
TLE2_HUMAN
|
||||||
| NC score | 0.015288 (rank : 143) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.014751 (rank : 144) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
TLE2_MOUSE
|
||||||
| NC score | 0.014637 (rank : 145) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVB2 | Gene names | Tle2, Grg2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2. | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.014417 (rank : 146) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
POLH_MOUSE
|
||||||
| NC score | 0.013370 (rank : 147) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJN0, Q9JJJ2 | Gene names | Polh, Rad30a, Xpv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein homolog). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.012774 (rank : 148) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.012533 (rank : 149) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
NPHP4_HUMAN
|
||||||
| NC score | 0.012397 (rank : 150) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75161 | Gene names | NPHP4, KIAA0673 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nephrocystin-4 (Nephroretinin). | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.012126 (rank : 151) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
IC1_MOUSE
|
||||||
| NC score | 0.011966 (rank : 152) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
PKHG1_HUMAN
|
||||||
| NC score | 0.011045 (rank : 153) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.010925 (rank : 154) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
PAX6_HUMAN
|
||||||
| NC score | 0.010406 (rank : 155) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26367, Q6N006, Q99413 | Gene names | PAX6, AN2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-6 (Oculorhombin) (Aniridia type II protein). | |||||
|
PAX6_MOUSE
|
||||||
| NC score | 0.010406 (rank : 156) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P63015, P32117, P70601, Q62222, Q64037, Q8CEI5, Q8VDB5, Q921Q8 | Gene names | Pax6, Pax-6, Sey | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-6 (Oculorhombin). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.010263 (rank : 157) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
C1QR1_HUMAN
|
||||||
| NC score | 0.009854 (rank : 158) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.009460 (rank : 159) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ARHGA_HUMAN
|
||||||
| NC score | 0.009097 (rank : 160) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.008411 (rank : 161) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MYPN_MOUSE
|
||||||
| NC score | 0.008354 (rank : 162) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
ZSWM4_MOUSE
|
||||||
| NC score | 0.008197 (rank : 163) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C7B8, Q8BPN9 | Gene names | Zswim4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger SWIM domain-containing protein 4. | |||||
|
MYL6B_HUMAN
|
||||||
| NC score | 0.008169 (rank : 164) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.007964 (rank : 165) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.007445 (rank : 166) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.006698 (rank : 167) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
ANKY2_MOUSE
|
||||||
| NC score | 0.006666 (rank : 168) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TPE9, Q8BK14, Q8BYW5, Q8R3N4, Q921J1 | Gene names | Ankmy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 2. | |||||
|
SOX7_HUMAN
|
||||||
| NC score | 0.006023 (rank : 169) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BT81 | Gene names | SOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7. | |||||
|
TRI37_MOUSE
|
||||||
| NC score | 0.005362 (rank : 170) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PCX9, Q8CHC5 | Gene names | Trim37, Kiaa0898 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37. | |||||
|
FOXN4_MOUSE
|
||||||
| NC score | 0.005221 (rank : 171) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Q3, Q920C0 | Gene names | Foxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N4. | |||||
|
NKX31_MOUSE
|
||||||
| NC score | 0.005110 (rank : 172) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97436, O09087 | Gene names | Nkx3-1, Nkx-3.1, Nkx3a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-3.1. | |||||
|
GBX2_HUMAN
|
||||||
| NC score | 0.004923 (rank : 173) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52951, O43833, Q53RX5, Q9Y5Y1 | Gene names | GBX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GBX-2 (Gastrulation and brain-specific homeobox protein 2). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | 0.004833 (rank : 174) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.004668 (rank : 175) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PCD12_HUMAN
|
||||||
| NC score | 0.004583 (rank : 176) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
FOXD3_HUMAN
|
||||||
| NC score | 0.003972 (rank : 177) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJU5, Q9BYM2, Q9UDD1 | Gene names | FOXD3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis). | |||||
|
HIRA_HUMAN
|
||||||
| NC score | 0.003907 (rank : 178) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54198, Q8IXN2 | Gene names | HIRA, DGCR1, HIR, TUPLE1 | |||
|
Domain Architecture |

|
|||||
| Description | HIRA protein (TUP1-like enhancer of split protein 1). | |||||
|
EGF_MOUSE
|
||||||
| NC score | 0.003471 (rank : 179) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.003321 (rank : 180) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.002227 (rank : 181) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BCL6_HUMAN
|
||||||
| NC score | 0.001457 (rank : 182) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41182 | Gene names | BCL6, BCL5, LAZ3, ZBTB27, ZNF51 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein (BCL-6) (Zinc finger protein 51) (LAZ-3 protein) (BCL-5) (Zinc finger and BTB domain-containing protein 27). | |||||
|
VGFR3_MOUSE
|
||||||
| NC score | -0.000368 (rank : 183) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35917 | Gene names | Flt4, Flt-4 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 3 precursor (EC 2.7.10.1) (VEGFR-3) (Tyrosine-protein kinase receptor FLT4). | |||||
|
SN1L1_MOUSE
|
||||||
| NC score | -0.000433 (rank : 184) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60670 | Gene names | Snf1lk, Msk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK) (HRT-20) (Myocardial SNF1- like kinase). | |||||
|
SNRK_HUMAN
|
||||||
| NC score | -0.000704 (rank : 185) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRH2, Q14706, Q68D15, Q6IQ46, Q9NXI7 | Gene names | SNRK, KIAA0096, SNFRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SNF-related serine/threonine-protein kinase (EC 2.7.11.1) (SNF1- related kinase). | |||||
|
ZFP95_HUMAN
|
||||||
| NC score | -0.000859 (rank : 186) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 144 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||