Please be patient as the page loads
|
TTLL5_MOUSE
|
||||||
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TTLL5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.974502 (rank : 2) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 150 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
TTLL4_HUMAN
|
||||||
| θ value | 3.22841e-82 (rank : 3) | NC score | 0.872267 (rank : 4) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TTLL4_MOUSE
|
||||||
| θ value | 2.0926e-81 (rank : 4) | NC score | 0.865674 (rank : 5) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TTLL2_HUMAN
|
||||||
| θ value | 1.55911e-60 (rank : 5) | NC score | 0.886997 (rank : 3) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BWV7, Q7Z6R8, Q86X22 | Gene names | TTLL2, C6orf104 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 2 (Testis-specific protein NYD- TSPG). | |||||
|
TTLL7_HUMAN
|
||||||
| θ value | 1.36585e-56 (rank : 6) | NC score | 0.861723 (rank : 6) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZT98, Q5TAX8, Q5TAX9, Q6P990, Q86YS1, Q9H5U4 | Gene names | TTLL7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 7 (Protein NYD-SP30). | |||||
|
TTLL1_HUMAN
|
||||||
| θ value | 8.02596e-49 (rank : 7) | NC score | 0.856557 (rank : 7) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
TTLL1_MOUSE
|
||||||
| θ value | 2.3352e-48 (rank : 8) | NC score | 0.856196 (rank : 8) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91V51, Q3TGC8, Q543S4, Q8C0A2, Q91ZG1 | Gene names | Ttll1 | |||
|
Domain Architecture |
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1) (p49). | |||||
|
TTL11_HUMAN
|
||||||
| θ value | 2.59387e-31 (rank : 9) | NC score | 0.833965 (rank : 9) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NHH1 | Gene names | TTLL11, C9orf20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 11. | |||||
|
TTLL3_HUMAN
|
||||||
| θ value | 5.23862e-24 (rank : 10) | NC score | 0.804675 (rank : 10) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y4R7, Q9UI99 | Gene names | TTLL3 | |||
|
Domain Architecture |
|
|||||
| Description | Tubulin--tyrosine ligase-like protein 3 (HOTTL). | |||||
|
TTL_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 11) | NC score | 0.685633 (rank : 11) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NG68, Q7Z302, Q8N426 | Gene names | TTL | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
TTL_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 12) | NC score | 0.684296 (rank : 12) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P38585, Q8R1L7, Q8VEG2 | Gene names | Ttl | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
TTL12_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 13) | NC score | 0.474777 (rank : 13) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14166, Q9UGU3 | Gene names | TTLL12, KIAA0153 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 12. | |||||
|
GP124_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.027743 (rank : 49) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PE1, Q8N3R1, Q8TEM3, Q96KB2, Q9P1Z7, Q9UFY4 | Gene names | GPR124, KIAA1531, TEM5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.036196 (rank : 29) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.066508 (rank : 14) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.041621 (rank : 25) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
NKX61_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.025654 (rank : 56) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78426 | Gene names | NKX6-1, NKX6A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
CI079_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.053380 (rank : 16) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
PHC2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.046196 (rank : 22) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.033713 (rank : 34) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.027267 (rank : 51) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
STK10_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.010698 (rank : 108) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TSSK2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.005210 (rank : 138) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PF2, Q8IY55 | Gene names | TSSK2, DGSG, SPOGA2, STK22B | |||
|
Domain Architecture |
|
|||||
| Description | Testis-specific serine/threonine-protein kinase 2 (EC 2.7.11.1) (TSSK- 2) (Testis-specific kinase 2) (TSK-2) (Serine/threonine-protein kinase 22B) (DiGeorge syndrome protein G). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.038693 (rank : 27) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.030665 (rank : 38) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.033627 (rank : 35) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.022362 (rank : 67) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
TRA2A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.024525 (rank : 63) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |
|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
AUXI_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.039842 (rank : 26) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
K0494_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.029010 (rank : 44) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75071 | Gene names | KIAA0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
NKX61_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.022487 (rank : 66) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.021006 (rank : 73) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.036846 (rank : 28) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
YYAP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.048602 (rank : 20) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H869, Q5VYZ4, Q5VYZ7, Q7L4C3, Q7L5E2, Q8IXA6, Q8TEW5, Q8TF04, Q96HB6, Q9BQ64, Q9NV84 | Gene names | YY1AP1, HCCA2, YY1AP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YY1-associated protein 1 (Hepatocellular carcinoma susceptibility protein) (Hepatocellular carcinoma-associated protein 2). | |||||
|
BARH2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.017142 (rank : 87) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VIB5, Q66L43 | Gene names | Barhl2, Barh1, Mbh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 2 homeobox protein (Bar-class homeodomain protein MBH1) (Homeobox protein B-H1). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.028327 (rank : 46) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
KLF17_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.004307 (rank : 140) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
MKL2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.025170 (rank : 58) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.050917 (rank : 19) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.035998 (rank : 30) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
AUXI_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.033280 (rank : 36) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.042613 (rank : 23) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.026668 (rank : 53) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.046254 (rank : 21) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.026651 (rank : 54) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
ZCH14_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.054854 (rank : 15) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.017273 (rank : 85) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.012453 (rank : 100) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.028283 (rank : 47) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.027708 (rank : 50) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.052364 (rank : 17) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.006615 (rank : 129) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.029907 (rank : 41) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CU058_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.051907 (rank : 18) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58505 | Gene names | C21orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf58. | |||||
|
ITPR3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.025901 (rank : 55) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.024942 (rank : 60) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.010562 (rank : 110) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.026682 (rank : 52) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.007029 (rank : 127) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
GP124_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.021707 (rank : 71) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
SIX2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.013733 (rank : 98) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62232, P70179 | Gene names | Six2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX2 (Sine oculis homeobox homolog 2). | |||||
|
TAF5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.005213 (rank : 137) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C092, Q8BTR2 | Gene names | Taf5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.011403 (rank : 104) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.008606 (rank : 117) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
CPSF7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.030274 (rank : 40) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
CPSF7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.030295 (rank : 39) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
GCX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.021901 (rank : 70) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.024402 (rank : 64) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NPIP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.034823 (rank : 32) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UND3 | Gene names | NPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex-interacting protein (NPIP). | |||||
|
REL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.016439 (rank : 92) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.021062 (rank : 72) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.028217 (rank : 48) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TRA2A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.018999 (rank : 80) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PFR5 | Gene names | Tra2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
UBN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.034057 (rank : 33) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
AAK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.008329 (rank : 121) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.012073 (rank : 102) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.022192 (rank : 68) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
GAH6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.022962 (rank : 65) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8A4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_22q11.2 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.017092 (rank : 88) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.028655 (rank : 45) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.029760 (rank : 43) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.035506 (rank : 31) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
PVRL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.013338 (rank : 99) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92692, O75455, Q96J29 | Gene names | PVRL2, HVEB, PRR2 | |||
|
Domain Architecture |
|
|||||
| Description | Poliovirus receptor-related protein 2 precursor (Herpes virus entry mediator B) (HveB) (Nectin-2) (CD112 antigen). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.009123 (rank : 115) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.020268 (rank : 77) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.011077 (rank : 105) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.007764 (rank : 122) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
CHP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.010675 (rank : 109) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
DCDC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.020418 (rank : 76) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.006549 (rank : 130) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.024646 (rank : 62) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
ITPR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.020606 (rank : 75) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
K2C1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.003004 (rank : 146) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04104 | Gene names | Krt1, Krt2-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.004007 (rank : 141) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MFN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.017305 (rank : 84) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U63, Q3V3B8, Q80WP4, Q80XK3, Q8BHF0, Q8BKV5, Q8R535, Q923X2 | Gene names | Mfn2, Kiaa0214, Marf | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN2 (EC 3.6.5.-) (Mitofusin-2) (Hypertension- related protein 1) (Mitochondrial assembly regulatory factor) (HSG protein). | |||||
|
NEUR4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.015081 (rank : 95) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWR8, Q96D64 | Gene names | NEU4 | |||
|
Domain Architecture |
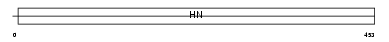
|
|||||
| Description | Sialidase-4 (EC 3.2.1.18) (N-acetyl-alpha-neuraminidase 4). | |||||
|
NUP98_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.025152 (rank : 59) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.005353 (rank : 136) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RANB9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.022154 (rank : 69) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
SP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.001687 (rank : 148) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.007388 (rank : 124) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.007082 (rank : 126) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
ITPR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.024886 (rank : 61) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.012166 (rank : 101) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.016668 (rank : 89) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RASA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.010108 (rank : 111) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.010925 (rank : 107) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
TEX9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.011052 (rank : 106) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.008564 (rank : 118) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.008446 (rank : 119) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.007133 (rank : 125) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.006060 (rank : 131) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.005918 (rank : 133) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.019045 (rank : 79) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.004531 (rank : 139) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.020766 (rank : 74) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.008335 (rank : 120) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.016661 (rank : 90) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.000826 (rank : 149) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.019380 (rank : 78) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.025186 (rank : 57) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.005546 (rank : 135) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
GLIS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | -0.000751 (rank : 150) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NEA6 | Gene names | GLIS3, ZNF515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3) (Zinc finger protein 515). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.018537 (rank : 83) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
HGS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.009467 (rank : 113) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.016598 (rank : 91) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
RASA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.009255 (rank : 114) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
TEX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.011876 (rank : 103) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.018682 (rank : 82) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
ZDH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.015377 (rank : 94) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8X9 | Gene names | ZDHHC11, ZNF399 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC11 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 11) (DHHC-11) (Zinc finger protein 399). | |||||
|
BLNK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.018761 (rank : 81) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.006020 (rank : 132) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.005855 (rank : 134) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CN152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.015547 (rank : 93) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N9Y4, Q96GY1 | Gene names | C14orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf152. | |||||
|
DICER_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.006713 (rank : 128) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.033022 (rank : 37) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.008810 (rank : 116) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.007581 (rank : 123) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.003357 (rank : 143) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
M3K4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.003098 (rank : 145) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.003201 (rank : 144) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MYOD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.029773 (rank : 42) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYPC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.001925 (rank : 147) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5XKE0, Q8C109, Q8K2V0 | Gene names | Mybpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.042233 (rank : 24) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
RANB9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.017160 (rank : 86) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.009660 (rank : 112) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.003363 (rank : 142) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.014762 (rank : 96) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.014342 (rank : 97) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 150 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.974502 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
TTLL2_HUMAN
|
||||||
| NC score | 0.886997 (rank : 3) | θ value | 1.55911e-60 (rank : 5) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BWV7, Q7Z6R8, Q86X22 | Gene names | TTLL2, C6orf104 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 2 (Testis-specific protein NYD- TSPG). | |||||
|
TTLL4_HUMAN
|
||||||
| NC score | 0.872267 (rank : 4) | θ value | 3.22841e-82 (rank : 3) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TTLL4_MOUSE
|
||||||
| NC score | 0.865674 (rank : 5) | θ value | 2.0926e-81 (rank : 4) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TTLL7_HUMAN
|
||||||
| NC score | 0.861723 (rank : 6) | θ value | 1.36585e-56 (rank : 6) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZT98, Q5TAX8, Q5TAX9, Q6P990, Q86YS1, Q9H5U4 | Gene names | TTLL7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 7 (Protein NYD-SP30). | |||||
|
TTLL1_HUMAN
|
||||||
| NC score | 0.856557 (rank : 7) | θ value | 8.02596e-49 (rank : 7) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
TTLL1_MOUSE
|
||||||
| NC score | 0.856196 (rank : 8) | θ value | 2.3352e-48 (rank : 8) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91V51, Q3TGC8, Q543S4, Q8C0A2, Q91ZG1 | Gene names | Ttll1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1) (p49). | |||||
|
TTL11_HUMAN
|
||||||
| NC score | 0.833965 (rank : 9) | θ value | 2.59387e-31 (rank : 9) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NHH1 | Gene names | TTLL11, C9orf20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 11. | |||||
|
TTLL3_HUMAN
|
||||||
| NC score | 0.804675 (rank : 10) | θ value | 5.23862e-24 (rank : 10) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y4R7, Q9UI99 | Gene names | TTLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 3 (HOTTL). | |||||
|
TTL_HUMAN
|
||||||
| NC score | 0.685633 (rank : 11) | θ value | 6.20254e-17 (rank : 11) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NG68, Q7Z302, Q8N426 | Gene names | TTL | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
TTL_MOUSE
|
||||||
| NC score | 0.684296 (rank : 12) | θ value | 1.38178e-16 (rank : 12) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P38585, Q8R1L7, Q8VEG2 | Gene names | Ttl | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
TTL12_HUMAN
|
||||||
| NC score | 0.474777 (rank : 13) | θ value | 8.97725e-08 (rank : 13) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14166, Q9UGU3 | Gene names | TTLL12, KIAA0153 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 12. | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.066508 (rank : 14) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ZCH14_HUMAN
|
||||||
| NC score | 0.054854 (rank : 15) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.053380 (rank : 16) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.052364 (rank : 17) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CU058_HUMAN
|
||||||
| NC score | 0.051907 (rank : 18) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58505 | Gene names | C21orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf58. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.050917 (rank : 19) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
YYAP1_HUMAN
|
||||||
| NC score | 0.048602 (rank : 20) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H869, Q5VYZ4, Q5VYZ7, Q7L4C3, Q7L5E2, Q8IXA6, Q8TEW5, Q8TF04, Q96HB6, Q9BQ64, Q9NV84 | Gene names | YY1AP1, HCCA2, YY1AP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YY1-associated protein 1 (Hepatocellular carcinoma susceptibility protein) (Hepatocellular carcinoma-associated protein 2). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.046254 (rank : 21) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PHC2_MOUSE
|
||||||
| NC score | 0.046196 (rank : 22) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.042613 (rank : 23) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.042233 (rank : 24) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.041621 (rank : 25) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
AUXI_HUMAN
|
||||||
| NC score | 0.039842 (rank : 26) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75061, Q32M66, Q4G0K1, Q5T614, Q5T615 | Gene names | DNAJC6, KIAA0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.038693 (rank : 27) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.036846 (rank : 28) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.036196 (rank : 29) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.035998 (rank : 30) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.035506 (rank : 31) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
NPIP_HUMAN
|
||||||
| NC score | 0.034823 (rank : 32) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UND3 | Gene names | NPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex-interacting protein (NPIP). | |||||
|
UBN1_HUMAN
|
||||||
| NC score | 0.034057 (rank : 33) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NPG3, Q13079, Q9P1P7 | Gene names | UBN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubinuclein (Ubiquitously expressed nuclear protein) (VT4). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.033713 (rank : 34) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.033627 (rank : 35) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
AUXI_MOUSE
|
||||||
| NC score | 0.033280 (rank : 36) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TZ3, Q6P2K9, Q8C7L9 | Gene names | Dnajc6, Kiaa0473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative tyrosine-protein phosphatase auxilin (EC 3.1.3.48) (DnaJ homolog subfamily C member 6). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.033022 (rank : 37) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.030665 (rank : 38) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
CPSF7_MOUSE
|
||||||
| NC score | 0.030295 (rank : 39) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BTV2, Q3TNF1, Q8BKE7, Q8CFS8 | Gene names | Cpsf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7. | |||||
|
CPSF7_HUMAN
|
||||||
| NC score | 0.030274 (rank : 40) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N684, Q7Z3H9, Q9H025, Q9H9V1 | Gene names | CPSF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 7 (Cleavage and polyadenylation specificity factor 59 kDa subunit) (CPSF 59 kDa subunit) (Pre-mRNA cleavage factor Im 59 kDa subunit). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.029907 (rank : 41) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
MYOD1_HUMAN
|
||||||
| NC score | 0.029773 (rank : 42) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.029760 (rank : 43) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
K0494_HUMAN
|
||||||
| NC score | 0.029010 (rank : 44) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75071 | Gene names | KIAA0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.028655 (rank : 45) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.028327 (rank : 46) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.028283 (rank : 47) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.028217 (rank : 48) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
GP124_HUMAN
|
||||||
| NC score | 0.027743 (rank : 49) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PE1, Q8N3R1, Q8TEM3, Q96KB2, Q9P1Z7, Q9UFY4 | Gene names | GPR124, KIAA1531, TEM5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.027708 (rank : 50) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.027267 (rank : 51) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.026682 (rank : 52) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.026668 (rank : 53) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.026651 (rank : 54) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
ITPR3_MOUSE
|
||||||
| NC score | 0.025901 (rank : 55) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
NKX61_HUMAN
|
||||||
| NC score | 0.025654 (rank : 56) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78426 | Gene names | NKX6-1, NKX6A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.025186 (rank : 57) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
MKL2_HUMAN
|
||||||
| NC score | 0.025170 (rank : 58) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.025152 (rank : 59) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.024942 (rank : 60) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ITPR2_MOUSE
|
||||||
| NC score | 0.024886 (rank : 61) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.024646 (rank : 62) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
TRA2A_HUMAN
|
||||||
| NC score | 0.024525 (rank : 63) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13595 | Gene names | TRA2A | |||
|
Domain Architecture |

|
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.024402 (rank : 64) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
GAH6_HUMAN
|
||||||
| NC score | 0.022962 (rank : 65) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8A4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_22q11.2 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
NKX61_MOUSE
|
||||||
| NC score | 0.022487 (rank : 66) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.022362 (rank : 67) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.022192 (rank : 68) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
RANB9_HUMAN
|
||||||
| NC score | 0.022154 (rank : 69) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
GCX1_HUMAN
|
||||||
| NC score | 0.021901 (rank : 70) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96NM4, Q5TE33, Q5TE34, Q5TE35, Q96IC9, Q9BQN5 | Gene names | GCX1, C20orf100 | |||
|
Domain Architecture |

|
|||||
| Description | Granulosa cell HMG box protein 1 (GCX-1). | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.021707 (rank : 71) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.021062 (rank : 72) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.021006 (rank : 73) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.020766 (rank : 74) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ITPR3_HUMAN
|
||||||
| NC score | 0.020606 (rank : 75) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14573, Q14649 | Gene names | ITPR3 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.020418 (rank : 76) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.020268 (rank : 77) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.019380 (rank : 78) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.019045 (rank : 79) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
TRA2A_MOUSE
|
||||||
| NC score | 0.018999 (rank : 80) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PFR5 | Gene names | Tra2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transformer-2 protein homolog (TRA-2 alpha). | |||||
|
BLNK_HUMAN
|
||||||
| NC score | 0.018761 (rank : 81) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.018682 (rank : 82) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.018537 (rank : 83) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MFN2_MOUSE
|
||||||
| NC score | 0.017305 (rank : 84) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U63, Q3V3B8, Q80WP4, Q80XK3, Q8BHF0, Q8BKV5, Q8R535, Q923X2 | Gene names | Mfn2, Kiaa0214, Marf | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN2 (EC 3.6.5.-) (Mitofusin-2) (Hypertension- related protein 1) (Mitochondrial assembly regulatory factor) (HSG protein). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.017273 (rank : 85) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
RANB9_MOUSE
|
||||||
| NC score | 0.017160 (rank : 86) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
BARH2_MOUSE
|
||||||
| NC score | 0.017142 (rank : 87) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VIB5, Q66L43 | Gene names | Barhl2, Barh1, Mbh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 2 homeobox protein (Bar-class homeodomain protein MBH1) (Homeobox protein B-H1). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.017092 (rank : 88) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.016668 (rank : 89) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.016661 (rank : 90) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.016598 (rank : 91) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.016439 (rank : 92) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
CN152_HUMAN
|
||||||
| NC score | 0.015547 (rank : 93) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N9Y4, Q96GY1 | Gene names | C14orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf152. | |||||
|
ZDH11_HUMAN
|
||||||
| NC score | 0.015377 (rank : 94) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8X9 | Gene names | ZDHHC11, ZNF399 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC11 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 11) (DHHC-11) (Zinc finger protein 399). | |||||
|
NEUR4_HUMAN
|
||||||
| NC score | 0.015081 (rank : 95) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWR8, Q96D64 | Gene names | NEU4 | |||
|
Domain Architecture |

|
|||||
| Description | Sialidase-4 (EC 3.2.1.18) (N-acetyl-alpha-neuraminidase 4). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.014762 (rank : 96) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.014342 (rank : 97) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
SIX2_MOUSE
|
||||||
| NC score | 0.013733 (rank : 98) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62232, P70179 | Gene names | Six2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX2 (Sine oculis homeobox homolog 2). | |||||
|
PVRL2_HUMAN
|
||||||
| NC score | 0.013338 (rank : 99) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92692, O75455, Q96J29 | Gene names | PVRL2, HVEB, PRR2 | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 2 precursor (Herpes virus entry mediator B) (HveB) (Nectin-2) (CD112 antigen). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.012453 (rank : 100) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.012166 (rank : 101) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.012073 (rank : 102) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TEX2_HUMAN
|
||||||
| NC score | 0.011876 (rank : 103) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.011403 (rank : 104) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.011077 (rank : 105) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
TEX9_MOUSE
|
||||||
| NC score | 0.011052 (rank : 106) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D845, O54764 | Gene names | Tex9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein (Testis-specifically expressed protein 1) (Tsec-1). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.010925 (rank : 107) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.010698 (rank : 108) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
CHP2_HUMAN
|
||||||
| NC score | 0.010675 (rank : 109) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.010562 (rank : 110) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
RASA3_HUMAN
|
||||||
| NC score | 0.010108 (rank : 111) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.009660 (rank : 112) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.009467 (rank : 113) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
RASA3_MOUSE
|
||||||
| NC score | 0.009255 (rank : 114) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.009123 (rank : 115) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.008810 (rank : 116) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.008606 (rank : 117) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.008564 (rank : 118) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.008446 (rank : 119) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.008335 (rank : 120) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.008329 (rank : 121) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.007764 (rank : 122) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.007581 (rank : 123) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.007388 (rank : 124) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.007133 (rank : 125) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.007082 (rank : 126) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.007029 (rank : 127) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.006713 (rank : 128) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.006615 (rank : 129) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.006549 (rank : 130) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.006060 (rank : 131) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.006020 (rank : 132) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.005918 (rank : 133) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.005855 (rank : 134) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.005546 (rank : 135) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.005353 (rank : 136) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
TAF5_MOUSE
|
||||||
| NC score | 0.005213 (rank : 137) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C092, Q8BTR2 | Gene names | Taf5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 5 (Transcription initiation factor TFIID 100 kDa subunit) (TAF(II)100) (TAFII-100) (TAFII100). | |||||
|
TSSK2_HUMAN
|
||||||
| NC score | 0.005210 (rank : 138) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96PF2, Q8IY55 | Gene names | TSSK2, DGSG, SPOGA2, STK22B | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific serine/threonine-protein kinase 2 (EC 2.7.11.1) (TSSK- 2) (Testis-specific kinase 2) (TSK-2) (Serine/threonine-protein kinase 22B) (DiGeorge syndrome protein G). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.004531 (rank : 139) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
KLF17_HUMAN
|
||||||
| NC score | 0.004307 (rank : 140) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.004007 (rank : 141) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.003363 (rank : 142) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.003357 (rank : 143) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.003201 (rank : 144) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
M3K4_MOUSE
|
||||||
| NC score | 0.003098 (rank : 145) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
K2C1_MOUSE
|
||||||
| NC score | 0.003004 (rank : 146) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04104 | Gene names | Krt1, Krt2-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin). | |||||
|
MYPC2_MOUSE
|
||||||
| NC score | 0.001925 (rank : 147) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5XKE0, Q8C109, Q8K2V0 | Gene names | Mybpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
SP2_HUMAN
|
||||||
| NC score | 0.001687 (rank : 148) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.000826 (rank : 149) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
GLIS3_HUMAN
|
||||||
| NC score | -0.000751 (rank : 150) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 150 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NEA6 | Gene names | GLIS3, ZNF515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3) (Zinc finger protein 515). | |||||