Please be patient as the page loads
|
TTLL4_MOUSE
|
||||||
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TTLL4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.980950 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TTLL4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 2.0926e-81 (rank : 3) | NC score | 0.865674 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 2.31364e-80 (rank : 4) | NC score | 0.871302 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
TTLL7_HUMAN
|
||||||
| θ value | 1.56274e-52 (rank : 5) | NC score | 0.860713 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZT98, Q5TAX8, Q5TAX9, Q6P990, Q86YS1, Q9H5U4 | Gene names | TTLL7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 7 (Protein NYD-SP30). | |||||
|
TTLL2_HUMAN
|
||||||
| θ value | 6.56568e-51 (rank : 6) | NC score | 0.876711 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BWV7, Q7Z6R8, Q86X22 | Gene names | TTLL2, C6orf104 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 2 (Testis-specific protein NYD- TSPG). | |||||
|
TTLL1_HUMAN
|
||||||
| θ value | 2.67181e-44 (rank : 7) | NC score | 0.853027 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |
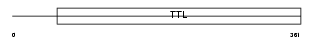
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
TTLL1_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 8) | NC score | 0.852637 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91V51, Q3TGC8, Q543S4, Q8C0A2, Q91ZG1 | Gene names | Ttll1 | |||
|
Domain Architecture |
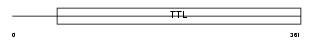
|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1) (p49). | |||||
|
TTL11_HUMAN
|
||||||
| θ value | 7.54701e-31 (rank : 9) | NC score | 0.838919 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHH1 | Gene names | TTLL11, C9orf20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 11. | |||||
|
TTLL3_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 10) | NC score | 0.789342 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y4R7, Q9UI99 | Gene names | TTLL3 | |||
|
Domain Architecture |
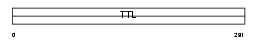
|
|||||
| Description | Tubulin--tyrosine ligase-like protein 3 (HOTTL). | |||||
|
TTL_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 11) | NC score | 0.694123 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NG68, Q7Z302, Q8N426 | Gene names | TTL | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
TTL_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 12) | NC score | 0.691312 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P38585, Q8R1L7, Q8VEG2 | Gene names | Ttl | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.048072 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TTL12_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 14) | NC score | 0.417476 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14166, Q9UGU3 | Gene names | TTLL12, KIAA0153 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 12. | |||||
|
ASPP2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 15) | NC score | 0.027507 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 16) | NC score | 0.064340 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.042724 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.024834 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
DCR1A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.041901 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.024524 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
IC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.019185 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05155, Q547W3, Q59EI5, Q96FE0 | Gene names | SERPING1, C1IN, C1NH | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.043242 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.039549 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.029443 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.045179 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
INHBA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.024908 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08476, Q14599 | Gene names | INHBA | |||
|
Domain Architecture |
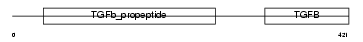
|
|||||
| Description | Inhibin beta A chain precursor (Activin beta-A chain) (Erythroid differentiation protein) (EDF). | |||||
|
INHBA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.025012 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04998 | Gene names | Inhba | |||
|
Domain Architecture |
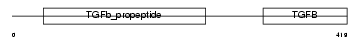
|
|||||
| Description | Inhibin beta A chain precursor (Activin beta-A chain). | |||||
|
MARK3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.005583 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.012945 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PHC2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.038169 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
MARK3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.005353 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
TRPS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.018396 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
YYAP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.040953 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H869, Q5VYZ4, Q5VYZ7, Q7L4C3, Q7L5E2, Q8IXA6, Q8TEW5, Q8TF04, Q96HB6, Q9BQ64, Q9NV84 | Gene names | YY1AP1, HCCA2, YY1AP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YY1-associated protein 1 (Hepatocellular carcinoma susceptibility protein) (Hepatocellular carcinoma-associated protein 2). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.009907 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
APC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.025814 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.009586 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BRSK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.004471 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
SYN2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.017969 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
ZN628_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | -0.000620 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
BRSK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.003887 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||
|
CCD82_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.027512 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.038926 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.017116 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.022284 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.011845 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.019298 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
FARP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.008700 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
FOXP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.007855 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
LRC41_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.041687 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1C9, Q91VX4 | Gene names | Lrrc41, Muf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.025819 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.019479 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.012444 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.015126 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PHC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.024259 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
PUS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.019115 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZE2, Q96D17, Q96J23, Q96NB4 | Gene names | PUS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA pseudouridine synthase 3 (EC 5.4.99.-) (tRNA-uridine isomerase 3) (tRNA pseudouridylate synthase 3). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.006805 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
BORG4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.017589 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JM96, Q3TNF3, Q9QZT8 | Gene names | Cdc42ep4, Borg4, Cep4 | |||
|
Domain Architecture |
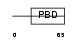
|
|||||
| Description | Cdc42 effector protein 4 (Binder of Rho GTPases 4). | |||||
|
DJC16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.008785 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
K0226_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.015704 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
LRC41_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.038947 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15345, Q71RA8, Q9BSM0 | Gene names | LRRC41, MUF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.030817 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SH24A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.018121 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
SNX15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.015190 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WE1 | Gene names | Snx15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-15. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.020038 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.012433 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
CCD82_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.024656 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PG04, Q3V462 | Gene names | Ccdc82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.002419 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.035083 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.020077 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
DSCAM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.004685 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60469, O60468 | Gene names | DSCAM | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome cell adhesion molecule precursor (CHD2). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.011555 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
IC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.013415 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
MCR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.004448 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.013232 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.015646 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ZDHC5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.007400 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |
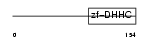
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.008118 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
ACF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.005714 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.026247 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.003695 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.013674 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
HLX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.004098 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14774, Q15988 | Gene names | HLX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HLX1 (Homeobox protein HB24). | |||||
|
HTF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.011727 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
HTF4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.012067 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.015191 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.016009 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PEX5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.009076 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.020014 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.020117 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.008422 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.038560 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.009990 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CEBPA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.008143 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
FZD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.003096 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60353, Q6P9C3 | Gene names | FZD6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (hFz6). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.006806 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
MAGB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.006109 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |
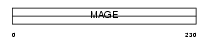
|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.017846 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.010293 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
MYLK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.000274 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.007674 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TLE4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.009830 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
ZN403_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.009712 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SV77, Q5SV75, Q5SV76, Q5SV78, Q6GVH6, Q6P9J4, Q920N4 | Gene names | Znf403, Ggnbp2, Zfp403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Gametogenetin-binding protein 2) (Dioxin-inducible factor 3) (DIF-3). | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.020454 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
TTLL4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TTLL4_HUMAN
|
||||||
| NC score | 0.980950 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TTLL2_HUMAN
|
||||||
| NC score | 0.876711 (rank : 3) | θ value | 6.56568e-51 (rank : 6) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BWV7, Q7Z6R8, Q86X22 | Gene names | TTLL2, C6orf104 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 2 (Testis-specific protein NYD- TSPG). | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.871302 (rank : 4) | θ value | 2.31364e-80 (rank : 4) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.865674 (rank : 5) | θ value | 2.0926e-81 (rank : 3) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
TTLL7_HUMAN
|
||||||
| NC score | 0.860713 (rank : 6) | θ value | 1.56274e-52 (rank : 5) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6ZT98, Q5TAX8, Q5TAX9, Q6P990, Q86YS1, Q9H5U4 | Gene names | TTLL7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 7 (Protein NYD-SP30). | |||||
|
TTLL1_HUMAN
|
||||||
| NC score | 0.853027 (rank : 7) | θ value | 2.67181e-44 (rank : 7) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95922, Q9BR27, Q9NRS9, Q9UMU0 | Gene names | TTLL1, C22orf7 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1). | |||||
|
TTLL1_MOUSE
|
||||||
| NC score | 0.852637 (rank : 8) | θ value | 3.48949e-44 (rank : 8) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91V51, Q3TGC8, Q543S4, Q8C0A2, Q91ZG1 | Gene names | Ttll1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable tubulin polyglutamylase (EC 6.-.-.-) (Tubulin polyglutamylase complex subunit 3) (PGs3) (Tubulin--tyrosine ligase-like protein 1) (p49). | |||||
|
TTL11_HUMAN
|
||||||
| NC score | 0.838919 (rank : 9) | θ value | 7.54701e-31 (rank : 9) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHH1 | Gene names | TTLL11, C9orf20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 11. | |||||
|
TTLL3_HUMAN
|
||||||
| NC score | 0.789342 (rank : 10) | θ value | 2.27762e-19 (rank : 10) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y4R7, Q9UI99 | Gene names | TTLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 3 (HOTTL). | |||||
|
TTL_HUMAN
|
||||||
| NC score | 0.694123 (rank : 11) | θ value | 2.13179e-17 (rank : 11) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NG68, Q7Z302, Q8N426 | Gene names | TTL | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
TTL_MOUSE
|
||||||
| NC score | 0.691312 (rank : 12) | θ value | 1.38178e-16 (rank : 12) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P38585, Q8R1L7, Q8VEG2 | Gene names | Ttl | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
TTL12_HUMAN
|
||||||
| NC score | 0.417476 (rank : 13) | θ value | 0.00298849 (rank : 14) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14166, Q9UGU3 | Gene names | TTLL12, KIAA0153 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 12. | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.064340 (rank : 14) | θ value | 0.0113563 (rank : 16) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.048072 (rank : 15) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.045179 (rank : 16) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.043242 (rank : 17) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.042724 (rank : 18) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
DCR1A_MOUSE
|
||||||
| NC score | 0.041901 (rank : 19) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
LRC41_MOUSE
|
||||||
| NC score | 0.041687 (rank : 20) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1C9, Q91VX4 | Gene names | Lrrc41, Muf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
YYAP1_HUMAN
|
||||||
| NC score | 0.040953 (rank : 21) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H869, Q5VYZ4, Q5VYZ7, Q7L4C3, Q7L5E2, Q8IXA6, Q8TEW5, Q8TF04, Q96HB6, Q9BQ64, Q9NV84 | Gene names | YY1AP1, HCCA2, YY1AP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YY1-associated protein 1 (Hepatocellular carcinoma susceptibility protein) (Hepatocellular carcinoma-associated protein 2). | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.039549 (rank : 22) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
LRC41_HUMAN
|
||||||
| NC score | 0.038947 (rank : 23) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15345, Q71RA8, Q9BSM0 | Gene names | LRRC41, MUF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.038926 (rank : 24) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.038560 (rank : 25) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
PHC2_MOUSE
|
||||||
| NC score | 0.038169 (rank : 26) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.035083 (rank : 27) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.030817 (rank : 28) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.029443 (rank : 29) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CCD82_HUMAN
|
||||||
| NC score | 0.027512 (rank : 30) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N4S0, Q9H2Q5, Q9H5E3 | Gene names | CCDC82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
ASPP2_HUMAN
|
||||||
| NC score | 0.027507 (rank : 31) | θ value | 0.00509761 (rank : 15) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13625, Q12892, Q86X75, Q96KQ3 | Gene names | TP53BP2, ASPP2, BBP | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 2 (Tumor suppressor p53-binding protein 2) (p53-binding protein 2) (p53BP2) (53BP2) (Bcl2-binding protein) (Bbp) (NY-REN-51 antigen). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.026247 (rank : 32) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.025819 (rank : 33) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.025814 (rank : 34) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
INHBA_MOUSE
|
||||||
| NC score | 0.025012 (rank : 35) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04998 | Gene names | Inhba | |||
|
Domain Architecture |

|
|||||
| Description | Inhibin beta A chain precursor (Activin beta-A chain). | |||||
|
INHBA_HUMAN
|
||||||
| NC score | 0.024908 (rank : 36) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08476, Q14599 | Gene names | INHBA | |||
|
Domain Architecture |

|
|||||
| Description | Inhibin beta A chain precursor (Activin beta-A chain) (Erythroid differentiation protein) (EDF). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.024834 (rank : 37) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
CCD82_MOUSE
|
||||||
| NC score | 0.024656 (rank : 38) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PG04, Q3V462 | Gene names | Ccdc82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 82. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.024524 (rank : 39) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PHC1_MOUSE
|
||||||
| NC score | 0.024259 (rank : 40) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.022284 (rank : 41) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.020454 (rank : 42) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.020117 (rank : 43) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.020077 (rank : 44) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.020038 (rank : 45) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.020014 (rank : 46) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.019479 (rank : 47) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.019298 (rank : 48) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
IC1_HUMAN
|
||||||
| NC score | 0.019185 (rank : 49) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05155, Q547W3, Q59EI5, Q96FE0 | Gene names | SERPING1, C1IN, C1NH | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
PUS3_HUMAN
|
||||||
| NC score | 0.019115 (rank : 50) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZE2, Q96D17, Q96J23, Q96NB4 | Gene names | PUS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA pseudouridine synthase 3 (EC 5.4.99.-) (tRNA-uridine isomerase 3) (tRNA pseudouridylate synthase 3). | |||||
|
TRPS1_HUMAN
|
||||||
| NC score | 0.018396 (rank : 51) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
SH24A_HUMAN
|
||||||
| NC score | 0.018121 (rank : 52) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
SYN2_MOUSE
|
||||||
| NC score | 0.017969 (rank : 53) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.017846 (rank : 54) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
BORG4_MOUSE
|
||||||
| NC score | 0.017589 (rank : 55) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JM96, Q3TNF3, Q9QZT8 | Gene names | Cdc42ep4, Borg4, Cep4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 4 (Binder of Rho GTPases 4). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.017116 (rank : 56) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.016009 (rank : 57) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
K0226_HUMAN
|
||||||
| NC score | 0.015704 (rank : 58) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.015646 (rank : 59) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.015191 (rank : 60) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
SNX15_MOUSE
|
||||||
| NC score | 0.015190 (rank : 61) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WE1 | Gene names | Snx15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-15. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.015126 (rank : 62) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.013674 (rank : 63) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
IC1_MOUSE
|
||||||
| NC score | 0.013415 (rank : 64) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97290, O88330, Q99M43, Q9QX09 | Gene names | Serping1, C1nh | |||
|
Domain Architecture |

|
|||||
| Description | Plasma protease C1 inhibitor precursor (C1 Inh) (C1Inh) (C1 esterase inhibitor) (C1-inhibiting factor). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.013232 (rank : 65) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.012945 (rank : 66) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.012444 (rank : 67) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.012433 (rank : 68) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
HTF4_MOUSE
|
||||||
| NC score | 0.012067 (rank : 69) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.011845 (rank : 70) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
HTF4_HUMAN
|
||||||
| NC score | 0.011727 (rank : 71) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.011555 (rank : 72) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.010293 (rank : 73) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.009990 (rank : 74) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.009907 (rank : 75) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
TLE4_HUMAN
|
||||||
| NC score | 0.009830 (rank : 76) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
ZN403_MOUSE
|
||||||
| NC score | 0.009712 (rank : 77) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SV77, Q5SV75, Q5SV76, Q5SV78, Q6GVH6, Q6P9J4, Q920N4 | Gene names | Znf403, Ggnbp2, Zfp403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Gametogenetin-binding protein 2) (Dioxin-inducible factor 3) (DIF-3). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.009586 (rank : 78) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
PEX5_HUMAN
|
||||||
| NC score | 0.009076 (rank : 79) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50542, Q15115, Q15266 | Gene names | PEX5, PXR1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal targeting signal 1 receptor (Peroxisome receptor 1) (Peroxisomal C-terminal targeting signal import receptor) (PTS1-BP) (Peroxin-5) (PTS1 receptor). | |||||
|
DJC16_MOUSE
|
||||||
| NC score | 0.008785 (rank : 80) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80TN4, Q811G1, Q8BHI2 | Gene names | Dnajc16, Kiaa0962 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 16 precursor. | |||||
|
FARP2_HUMAN
|
||||||
| NC score | 0.008700 (rank : 81) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94887, Q53QM5, Q8WU27, Q9UFE7 | Gene names | FARP2, KIAA0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.008422 (rank : 82) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.008143 (rank : 83) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.008118 (rank : 84) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
FOXP4_HUMAN
|
||||||
| NC score | 0.007855 (rank : 85) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVH2, Q96E19 | Gene names | FOXP4, FKHLA | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein P4 (Fork head-related protein-like A). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.007674 (rank : 86) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
ZDHC5_MOUSE
|
||||||
| NC score | 0.007400 (rank : 87) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.006806 (rank : 88) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.006805 (rank : 89) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
MAGB3_HUMAN
|
||||||
| NC score | 0.006109 (rank : 90) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15480, O75861 | Gene names | MAGEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B3 (MAGE-B3 antigen). | |||||
|
ACF_MOUSE
|
||||||
| NC score | 0.005714 (rank : 91) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
MARK3_MOUSE
|
||||||
| NC score | 0.005583 (rank : 92) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
MARK3_HUMAN
|
||||||
| NC score | 0.005353 (rank : 93) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27448, O60219, Q8TB41, Q8WX83, Q96RG1, Q9UMY9, Q9UN34 | Gene names | MARK3, CTAK1 | |||
|
Domain Architecture |

|
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (Cdc25C- associated protein kinase 1) (cTAK1) (C-TAK1) (Serine/threonine protein kinase p78) (Ser/Thr protein kinase PAR-1) (Protein kinase STK10). | |||||
|
DSCAM_HUMAN
|
||||||
| NC score | 0.004685 (rank : 94) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60469, O60468 | Gene names | DSCAM | |||
|
Domain Architecture |

|
|||||
| Description | Down syndrome cell adhesion molecule precursor (CHD2). | |||||
|
BRSK2_HUMAN
|
||||||
| NC score | 0.004471 (rank : 95) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
MCR_MOUSE
|
||||||
| NC score | 0.004448 (rank : 96) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
HLX1_HUMAN
|
||||||
| NC score | 0.004098 (rank : 97) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14774, Q15988 | Gene names | HLX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HLX1 (Homeobox protein HB24). | |||||
|
BRSK1_HUMAN
|
||||||
| NC score | 0.003887 (rank : 98) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.003695 (rank : 99) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
FZD6_HUMAN
|
||||||
| NC score | 0.003096 (rank : 100) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60353, Q6P9C3 | Gene names | FZD6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (hFz6). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.002419 (rank : 101) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
MYLK_HUMAN
|
||||||
| NC score | 0.000274 (rank : 102) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
ZN628_HUMAN
|
||||||
| NC score | -0.000620 (rank : 103) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 103 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||