Please be patient as the page loads
|
CAC1E_MOUSE
|
||||||
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.968489 (rank : 6) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.977841 (rank : 3) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.976469 (rank : 4) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.973342 (rank : 5) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.956957 (rank : 13) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.958504 (rank : 12) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.959422 (rank : 11) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.997796 (rank : 2) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 167 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.960243 (rank : 10) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.960455 (rank : 9) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.961648 (rank : 8) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.962186 (rank : 7) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 4.83544e-70 (rank : 14) | NC score | 0.856358 (rank : 25) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 4.83544e-70 (rank : 15) | NC score | 0.856030 (rank : 27) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 6.31527e-70 (rank : 16) | NC score | 0.865661 (rank : 20) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 4.09345e-69 (rank : 17) | NC score | 0.855745 (rank : 28) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 9.11921e-69 (rank : 18) | NC score | 0.863495 (rank : 21) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 9.11921e-69 (rank : 19) | NC score | 0.870185 (rank : 18) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 1.5555e-68 (rank : 20) | NC score | 0.858264 (rank : 23) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 2.03155e-68 (rank : 21) | NC score | 0.857750 (rank : 24) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 2.03155e-68 (rank : 22) | NC score | 0.856352 (rank : 26) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 2.03155e-68 (rank : 23) | NC score | 0.854025 (rank : 29) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 2.65329e-68 (rank : 24) | NC score | 0.881842 (rank : 15) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 2.65329e-68 (rank : 25) | NC score | 0.904291 (rank : 14) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 5.91091e-68 (rank : 26) | NC score | 0.877552 (rank : 16) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 9.43692e-66 (rank : 27) | NC score | 0.871853 (rank : 17) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 1.2325e-65 (rank : 28) | NC score | 0.867231 (rank : 19) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 3.58603e-65 (rank : 29) | NC score | 0.853254 (rank : 30) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 8.55514e-59 (rank : 30) | NC score | 0.862984 (rank : 22) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 31) | NC score | 0.347024 (rank : 34) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 32) | NC score | 0.382014 (rank : 31) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 33) | NC score | 0.349167 (rank : 33) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 34) | NC score | 0.355071 (rank : 32) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 35) | NC score | 0.332170 (rank : 35) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 36) | NC score | 0.183720 (rank : 46) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 37) | NC score | 0.183898 (rank : 45) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 38) | NC score | 0.189952 (rank : 42) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 39) | NC score | 0.190019 (rank : 41) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 40) | NC score | 0.226747 (rank : 37) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 41) | NC score | 0.236226 (rank : 36) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 42) | NC score | 0.171625 (rank : 47) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 43) | NC score | 0.167737 (rank : 48) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 44) | NC score | 0.201963 (rank : 39) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 45) | NC score | 0.197505 (rank : 40) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 46) | NC score | 0.043712 (rank : 92) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 47) | NC score | 0.083221 (rank : 65) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 48) | NC score | 0.039503 (rank : 96) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 49) | NC score | 0.009221 (rank : 127) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 50) | NC score | 0.203732 (rank : 38) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 51) | NC score | 0.061021 (rank : 80) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 52) | NC score | 0.045257 (rank : 91) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
GPR63_MOUSE
|
||||||
| θ value | 0.163984 (rank : 53) | NC score | 0.007406 (rank : 134) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQQ3 | Gene names | Gpr63 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 63 (PSP24-beta) (PSP24-2). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 54) | NC score | 0.029388 (rank : 100) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.143630 (rank : 50) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
DMN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.009032 (rank : 128) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.121559 (rank : 56) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.108753 (rank : 59) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 0.21417 (rank : 59) | NC score | 0.027208 (rank : 101) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TCP10_HUMAN
|
||||||
| θ value | 0.21417 (rank : 60) | NC score | 0.041315 (rank : 95) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12799 | Gene names | TCP10, TCP10A | |||
|
Domain Architecture |

|
|||||
| Description | T-complex protein 10A homolog. | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 61) | NC score | 0.055364 (rank : 86) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 62) | NC score | 0.016331 (rank : 105) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
ULK2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 63) | NC score | -0.001333 (rank : 186) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 64) | NC score | 0.189109 (rank : 44) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 65) | NC score | 0.189142 (rank : 43) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 66) | NC score | 0.033870 (rank : 97) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
CENPC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 67) | NC score | 0.031534 (rank : 98) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 68) | NC score | 0.005149 (rank : 159) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.128732 (rank : 54) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.135337 (rank : 52) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 71) | NC score | 0.003219 (rank : 170) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
GPR63_HUMAN
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.004972 (rank : 160) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZJ6, Q9UJH3 | Gene names | GPR63, PSP24B | |||
|
Domain Architecture |
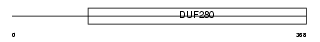
|
|||||
| Description | Probable G-protein coupled receptor 63 (PSP24-beta) (PSP24-2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.133354 (rank : 53) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 74) | NC score | 0.105473 (rank : 61) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
SN1L2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | -0.002226 (rank : 191) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.015381 (rank : 107) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.015352 (rank : 108) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.051499 (rank : 87) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SN1L2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | -0.002522 (rank : 192) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.012199 (rank : 118) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.027105 (rank : 102) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.008056 (rank : 131) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
OVGP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.006824 (rank : 139) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62010 | Gene names | Ovgp1, Chit5, Ogp | |||
|
Domain Architecture |

|
|||||
| Description | Oviduct-specific glycoprotein precursor (Oviductal glycoprotein) (Oviductin) (Estrogen-dependent oviduct protein). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.001178 (rank : 176) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SRTD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.013568 (rank : 112) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
TPTE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.100221 (rank : 62) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.041416 (rank : 94) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.020670 (rank : 104) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.010953 (rank : 120) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.083806 (rank : 64) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
SORC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.005602 (rank : 154) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.042916 (rank : 93) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.026460 (rank : 103) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
BOREA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.012567 (rank : 114) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q53HL2, Q53HN1, Q96AM3, Q9NVW5 | Gene names | CDCA8, PESCRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Borealin (Dasra-B) (hDasra-B) (Cell division cycle-associated protein 8) (Pluripotent embryonic stem cell-related gene 3 protein). | |||||
|
DCJ14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.007772 (rank : 132) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
JIP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.014705 (rank : 109) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
TRI55_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.005420 (rank : 155) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |
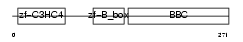
|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.013864 (rank : 110) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CHFR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.006220 (rank : 144) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EP1, Q96SL3, Q9NRT4, Q9NT32, Q9NVD5 | Gene names | CHFR | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
CT174_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.013735 (rank : 111) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
DHX36_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.006410 (rank : 142) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
IKKE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | -0.001790 (rank : 189) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0T8 | Gene names | Ikbke, Ikke, Ikki | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa-B kinase epsilon subunit (EC 2.7.11.10) (I kappa-B kinase epsilon) (IkBKE) (IKK-epsilon) (IKK- E) (Inducible I kappa-B kinase) (IKK-i). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.167314 (rank : 49) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
MBN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.010133 (rank : 123) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.007112 (rank : 136) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
RB15B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.015865 (rank : 106) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SON_MOUSE
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.012564 (rank : 115) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TBC10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.006987 (rank : 138) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58802 | Gene names | Tbc1d10a, Epi64, Tbc1d10 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
VN1B8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.005348 (rank : 157) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQ45 | Gene names | V1rb8, V1ra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-1 receptor B8 (Vomeronasal type-1 receptor A13). | |||||
|
AK1A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.006540 (rank : 140) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14550 | Gene names | AKR1A1, ALDR1, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Alcohol dehydrogenase [NADP+] (EC 1.1.1.2) (Aldehyde reductase) (Aldo- keto reductase family 1 member A1). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | -0.001855 (rank : 190) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.012228 (rank : 117) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.007549 (rank : 133) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
OSBL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.003795 (rank : 166) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.009431 (rank : 125) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.001111 (rank : 177) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SIAL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.010222 (rank : 122) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61711, Q61363 | Gene names | Ibsp | |||
|
Domain Architecture |
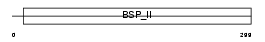
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.005738 (rank : 152) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.005641 (rank : 153) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.005988 (rank : 146) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.004392 (rank : 165) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TSN17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.006269 (rank : 143) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96FV3, Q96S98, Q9UKB9 | Gene names | TSPAN17, FBXO23, TM4SF17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetraspanin-17 (Tspan-17) (Transmembrane 4 superfamily member 17) (Tetraspan protein SB134) (F-box only protein 23). | |||||
|
TSN17_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.006459 (rank : 141) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7W4, Q91VI6 | Gene names | Tspan17, Fbxo23, Tm4sf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetraspanin-17 (Tspan-17) (Transmembrane 4 superfamily member 17) (Tetraspan protein SB134) (F-box only protein 23). | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.003208 (rank : 171) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.003730 (rank : 167) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
UTX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.005150 (rank : 158) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | -0.001153 (rank : 185) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.006215 (rank : 145) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.009532 (rank : 124) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.005854 (rank : 147) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
EVPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.001055 (rank : 178) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
GP73_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.005753 (rank : 151) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NBJ4, Q6IAF4, Q9NRB9 | Gene names | GOLPH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.059409 (rank : 81) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
MMRN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.004600 (rank : 164) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H8L6, Q6P2N2 | Gene names | MMRN2, EMILIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-2 precursor (EMILIN-3) (Elastin microfibril interface located protein 3) (Elastin microfibril interfacer 3) (EndoGlyx-1 p125/p140 subunit). | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.107262 (rank : 60) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.011439 (rank : 119) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.029800 (rank : 99) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TLR8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.000475 (rank : 179) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58682, Q91XI7 | Gene names | Tlr8 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 8 precursor. | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.005833 (rank : 149) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
TTLL4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.003695 (rank : 168) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.012792 (rank : 113) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.004707 (rank : 162) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
AK1A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.004927 (rank : 161) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JII6, Q9CQI5, Q9CQT8, Q9CT53, Q9D012, Q9D016, Q9D0I7, Q9D0P3 | Gene names | Akr1a1, Akr1a4 | |||
|
Domain Architecture |

|
|||||
| Description | Alcohol dehydrogenase [NADP+] (EC 1.1.1.2) (Aldehyde reductase) (Aldo- keto reductase family 1 member A1). | |||||
|
CI078_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.009242 (rank : 126) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
|
CI078_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.008349 (rank : 130) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TQI7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 homolog. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | -0.001345 (rank : 187) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.005808 (rank : 150) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.007154 (rank : 135) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.001506 (rank : 175) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
F120C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.005836 (rank : 148) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C3F2 | Gene names | Fam120c, ORF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C. | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.000240 (rank : 180) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.000073 (rank : 182) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.008976 (rank : 129) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.012407 (rank : 116) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.010659 (rank : 121) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.141710 (rank : 51) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
LAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | -0.001449 (rank : 188) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.002204 (rank : 174) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.002687 (rank : 173) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
NTBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.003221 (rank : 169) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.004627 (rank : 163) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | -0.000103 (rank : 183) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | -0.000315 (rank : 184) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.003195 (rank : 172) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.000116 (rank : 181) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
TTC28_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.007042 (rank : 137) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.005405 (rank : 156) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.082074 (rank : 66) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.083932 (rank : 63) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.075585 (rank : 70) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.076465 (rank : 68) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.067380 (rank : 76) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.066411 (rank : 77) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.056464 (rank : 84) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.056821 (rank : 83) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.064407 (rank : 78) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.069995 (rank : 73) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.072862 (rank : 71) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.078381 (rank : 67) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.110952 (rank : 57) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.109356 (rank : 58) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.070689 (rank : 72) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.068113 (rank : 75) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.063421 (rank : 79) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.057649 (rank : 82) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.069448 (rank : 74) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.050074 (rank : 90) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.051159 (rank : 89) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.076409 (rank : 69) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.125857 (rank : 55) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
MCLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.051489 (rank : 88) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.055774 (rank : 85) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 167 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.997796 (rank : 2) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.977841 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.976469 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.973342 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.968489 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.962186 (rank : 7) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.961648 (rank : 8) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.960455 (rank : 9) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.960243 (rank : 10) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.959422 (rank : 11) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.958504 (rank : 12) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.956957 (rank : 13) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.904291 (rank : 14) | θ value | 2.65329e-68 (rank : 25) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.881842 (rank : 15) | θ value | 2.65329e-68 (rank : 24) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.877552 (rank : 16) | θ value | 5.91091e-68 (rank : 26) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.871853 (rank : 17) | θ value | 9.43692e-66 (rank : 27) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.870185 (rank : 18) | θ value | 9.11921e-69 (rank : 19) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.867231 (rank : 19) | θ value | 1.2325e-65 (rank : 28) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.865661 (rank : 20) | θ value | 6.31527e-70 (rank : 16) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.863495 (rank : 21) | θ value | 9.11921e-69 (rank : 18) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.862984 (rank : 22) | θ value | 8.55514e-59 (rank : 30) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.858264 (rank : 23) | θ value | 1.5555e-68 (rank : 20) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.857750 (rank : 24) | θ value | 2.03155e-68 (rank : 21) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.856358 (rank : 25) | θ value | 4.83544e-70 (rank : 14) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.856352 (rank : 26) | θ value | 2.03155e-68 (rank : 22) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.856030 (rank : 27) | θ value | 4.83544e-70 (rank : 15) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.855745 (rank : 28) | θ value | 4.09345e-69 (rank : 17) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.854025 (rank : 29) | θ value | 2.03155e-68 (rank : 23) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.853254 (rank : 30) | θ value | 3.58603e-65 (rank : 29) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.382014 (rank : 31) | θ value | 2.21117e-06 (rank : 32) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.355071 (rank : 32) | θ value | 1.87187e-05 (rank : 34) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.349167 (rank : 33) | θ value | 4.92598e-06 (rank : 33) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.347024 (rank : 34) | θ value | 3.08544e-08 (rank : 31) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.332170 (rank : 35) | θ value | 1.87187e-05 (rank : 35) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.236226 (rank : 36) | θ value | 0.000786445 (rank : 41) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.226747 (rank : 37) | θ value | 0.000602161 (rank : 40) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.203732 (rank : 38) | θ value | 0.0736092 (rank : 50) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.201963 (rank : 39) | θ value | 0.00390308 (rank : 44) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.197505 (rank : 40) | θ value | 0.00869519 (rank : 45) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.190019 (rank : 41) | θ value | 3.19293e-05 (rank : 39) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.189952 (rank : 42) | θ value | 3.19293e-05 (rank : 38) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.189142 (rank : 43) | θ value | 0.365318 (rank : 65) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.189109 (rank : 44) | θ value | 0.365318 (rank : 64) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.183898 (rank : 45) | θ value | 3.19293e-05 (rank : 37) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.183720 (rank : 46) | θ value | 3.19293e-05 (rank : 36) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.171625 (rank : 47) | θ value | 0.00134147 (rank : 42) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.167737 (rank : 48) | θ value | 0.00175202 (rank : 43) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.167314 (rank : 49) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.143630 (rank : 50) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.141710 (rank : 51) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.135337 (rank : 52) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.133354 (rank : 53) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.128732 (rank : 54) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.125857 (rank : 55) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.121559 (rank : 56) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.110952 (rank : 57) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.109356 (rank : 58) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.108753 (rank : 59) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.107262 (rank : 60) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.105473 (rank : 61) | θ value | 0.813845 (rank : 74) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.100221 (rank : 62) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.083932 (rank : 63) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.083806 (rank : 64) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.083221 (rank : 65) | θ value | 0.0193708 (rank : 47) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.082074 (rank : 66) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.078381 (rank : 67) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.076465 (rank : 68) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.076409 (rank : 69) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.075585 (rank : 70) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.072862 (rank : 71) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.070689 (rank : 72) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.069995 (rank : 73) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.069448 (rank : 74) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.068113 (rank : 75) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.067380 (rank : 76) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.066411 (rank : 77) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.064407 (rank : 78) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.063421 (rank : 79) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.061021 (rank : 80) | θ value | 0.0736092 (rank : 51) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.059409 (rank : 81) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.057649 (rank : 82) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.056821 (rank : 83) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.056464 (rank : 84) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.055774 (rank : 85) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.055364 (rank : 86) | θ value | 0.21417 (rank : 61) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.051499 (rank : 87) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MCLN2_HUMAN
|
||||||
| NC score | 0.051489 (rank : 88) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.051159 (rank : 89) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.050074 (rank : 90) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.045257 (rank : 91) | θ value | 0.0961366 (rank : 52) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.043712 (rank : 92) | θ value | 0.00869519 (rank : 46) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.042916 (rank : 93) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.041416 (rank : 94) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TCP10_HUMAN
|
||||||
| NC score | 0.041315 (rank : 95) | θ value | 0.21417 (rank : 60) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12799 | Gene names | TCP10, TCP10A | |||
|
Domain Architecture |

|
|||||
| Description | T-complex protein 10A homolog. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.039503 (rank : 96) | θ value | 0.0193708 (rank : 48) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.033870 (rank : 97) | θ value | 0.365318 (rank : 66) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
CENPC_MOUSE
|
||||||
| NC score | 0.031534 (rank : 98) | θ value | 0.47712 (rank : 67) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.029800 (rank : 99) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.029388 (rank : 100) | θ value | 0.163984 (rank : 54) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.027208 (rank : 101) | θ value | 0.21417 (rank : 59) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.027105 (rank : 102) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.026460 (rank : 103) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.020670 (rank : 104) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.016331 (rank : 105) | θ value | 0.279714 (rank : 62) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.015865 (rank : 106) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.015381 (rank : 107) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.015352 (rank : 108) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.014705 (rank : 109) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.013864 (rank : 110) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.013735 (rank : 111) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
SRTD2_HUMAN
|
||||||
| NC score | 0.013568 (rank : 112) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.012792 (rank : 113) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
BOREA_HUMAN
|
||||||
| NC score | 0.012567 (rank : 114) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q53HL2, Q53HN1, Q96AM3, Q9NVW5 | Gene names | CDCA8, PESCRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Borealin (Dasra-B) (hDasra-B) (Cell division cycle-associated protein 8) (Pluripotent embryonic stem cell-related gene 3 protein). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.012564 (rank : 115) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.012407 (rank : 116) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.012228 (rank : 117) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.012199 (rank : 118) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.011439 (rank : 119) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.010953 (rank : 120) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.010659 (rank : 121) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
SIAL_MOUSE
|
||||||
| NC score | 0.010222 (rank : 122) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61711, Q61363 | Gene names | Ibsp | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
MBN3_MOUSE
|
||||||
| NC score | 0.010133 (rank : 123) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.009532 (rank : 124) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.009431 (rank : 125) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
CI078_HUMAN
|
||||||
| NC score | 0.009242 (rank : 126) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZ63, Q8WVU6, Q9NT39 | Gene names | C9orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 (Hepatocellular carcinoma-associated antigen 59). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.009221 (rank : 127) | θ value | 0.0736092 (rank : 49) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
DMN_HUMAN
|
||||||
| NC score | 0.009032 (rank : 128) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.008976 (rank : 129) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
CI078_MOUSE
|
||||||
| NC score | 0.008349 (rank : 130) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TQI7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf78 homolog. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.008056 (rank : 131) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
DCJ14_HUMAN
|
||||||
| NC score | 0.007772 (rank : 132) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6Y2X3, Q66K17, Q96N59, Q96T63 | Gene names | DNAJC14, DRIP78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14 (Dopamine receptor-interacting protein of 78 kDa) (DRiP78). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.007549 (rank : 133) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
GPR63_MOUSE
|
||||||
| NC score | 0.007406 (rank : 134) | θ value | 0.163984 (rank : 53) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQQ3 | Gene names | Gpr63 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 63 (PSP24-beta) (PSP24-2). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.007154 (rank : 135) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.007112 (rank : 136) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
TTC28_HUMAN
|
||||||
| NC score | 0.007042 (rank : 137) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96AY4, O95928, O95929, Q9NTE4, Q9UG31, Q9UGG5, Q9UPV8, Q9Y3S5 | Gene names | TTC28, KIAA1043 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
TBC10_MOUSE
|
||||||
| NC score | 0.006987 (rank : 138) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58802 | Gene names | Tbc1d10a, Epi64, Tbc1d10 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
OVGP1_MOUSE
|
||||||
| NC score | 0.006824 (rank : 139) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62010 | Gene names | Ovgp1, Chit5, Ogp | |||
|
Domain Architecture |

|
|||||
| Description | Oviduct-specific glycoprotein precursor (Oviductal glycoprotein) (Oviductin) (Estrogen-dependent oviduct protein). | |||||
|
AK1A1_HUMAN
|
||||||
| NC score | 0.006540 (rank : 140) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14550 | Gene names | AKR1A1, ALDR1, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Alcohol dehydrogenase [NADP+] (EC 1.1.1.2) (Aldehyde reductase) (Aldo- keto reductase family 1 member A1). | |||||
|
TSN17_MOUSE
|
||||||
| NC score | 0.006459 (rank : 141) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D7W4, Q91VI6 | Gene names | Tspan17, Fbxo23, Tm4sf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetraspanin-17 (Tspan-17) (Transmembrane 4 superfamily member 17) (Tetraspan protein SB134) (F-box only protein 23). | |||||
|
DHX36_HUMAN
|
||||||
| NC score | 0.006410 (rank : 142) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
TSN17_HUMAN
|
||||||
| NC score | 0.006269 (rank : 143) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96FV3, Q96S98, Q9UKB9 | Gene names | TSPAN17, FBXO23, TM4SF17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetraspanin-17 (Tspan-17) (Transmembrane 4 superfamily member 17) (Tetraspan protein SB134) (F-box only protein 23). | |||||
|
CHFR_HUMAN
|
||||||
| NC score | 0.006220 (rank : 144) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EP1, Q96SL3, Q9NRT4, Q9NT32, Q9NVD5 | Gene names | CHFR | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.006215 (rank : 145) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.005988 (rank : 146) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.005854 (rank : 147) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
F120C_MOUSE
|
||||||
| NC score | 0.005836 (rank : 148) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C3F2 | Gene names | Fam120c, ORF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C. | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.005833 (rank : 149) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.005808 (rank : 150) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
GP73_HUMAN
|
||||||
| NC score | 0.005753 (rank : 151) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NBJ4, Q6IAF4, Q9NRB9 | Gene names | GOLPH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi phosphoprotein 2 (Golgi membrane protein GP73). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.005738 (rank : 152) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.005641 (rank : 153) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SORC3_HUMAN
|
||||||
| NC score | 0.005602 (rank : 154) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
TRI55_HUMAN
|
||||||
| NC score | 0.005420 (rank : 155) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.005405 (rank : 156) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
VN1B8_MOUSE
|
||||||
| NC score | 0.005348 (rank : 157) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQ45 | Gene names | V1rb8, V1ra13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-1 receptor B8 (Vomeronasal type-1 receptor A13). | |||||
|
UTX_HUMAN
|
||||||
| NC score | 0.005150 (rank : 158) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.005149 (rank : 159) | θ value | 0.47712 (rank : 68) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
GPR63_HUMAN
|
||||||
| NC score | 0.004972 (rank : 160) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZJ6, Q9UJH3 | Gene names | GPR63, PSP24B | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 63 (PSP24-beta) (PSP24-2). | |||||
|
AK1A1_MOUSE
|
||||||
| NC score | 0.004927 (rank : 161) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JII6, Q9CQI5, Q9CQT8, Q9CT53, Q9D012, Q9D016, Q9D0I7, Q9D0P3 | Gene names | Akr1a1, Akr1a4 | |||
|
Domain Architecture |

|
|||||
| Description | Alcohol dehydrogenase [NADP+] (EC 1.1.1.2) (Aldehyde reductase) (Aldo- keto reductase family 1 member A1). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.004707 (rank : 162) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.004627 (rank : 163) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
MMRN2_HUMAN
|
||||||
| NC score | 0.004600 (rank : 164) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H8L6, Q6P2N2 | Gene names | MMRN2, EMILIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-2 precursor (EMILIN-3) (Elastin microfibril interface located protein 3) (Elastin microfibril interfacer 3) (EndoGlyx-1 p125/p140 subunit). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.004392 (rank : 165) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
OSBL5_HUMAN
|
||||||
| NC score | 0.003795 (rank : 166) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0X9, Q9BZB0, Q9P1Z4 | Gene names | OSBPL5, KIAA1534, ORP5 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.003730 (rank : 167) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
TTLL4_MOUSE
|
||||||
| NC score | 0.003695 (rank : 168) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80UG8, Q5U4C4 | Gene names | Ttll4, Kiaa0173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
NTBP1_MOUSE
|
||||||
| NC score | 0.003221 (rank : 169) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.003219 (rank : 170) | θ value | 0.62314 (rank : 71) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.003208 (rank : 171) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.003195 (rank : 172) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.002687 (rank : 173) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.002204 (rank : 174) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.001506 (rank : 175) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.001178 (rank : 176) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.001111 (rank : 177) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.001055 (rank : 178) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
TLR8_MOUSE
|
||||||
| NC score | 0.000475 (rank : 179) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58682, Q91XI7 | Gene names | Tlr8 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 8 precursor. | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.000240 (rank : 180) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.000116 (rank : 181) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.000073 (rank : 182) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | -0.000103 (rank : 183) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | -0.000315 (rank : 184) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | -0.001153 (rank : 185) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ULK2_MOUSE
|
||||||
| NC score | -0.001333 (rank : 186) | θ value | 0.279714 (rank : 63) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
CP250_HUMAN
|
||||||
| NC score | -0.001345 (rank : 187) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
LAP2_HUMAN
|
||||||
| NC score | -0.001449 (rank : 188) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
IKKE_MOUSE
|
||||||
| NC score | -0.001790 (rank : 189) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0T8 | Gene names | Ikbke, Ikke, Ikki | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa-B kinase epsilon subunit (EC 2.7.11.10) (I kappa-B kinase epsilon) (IkBKE) (IKK-epsilon) (IKK- E) (Inducible I kappa-B kinase) (IKK-i). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | -0.001855 (rank : 190) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
SN1L2_HUMAN
|
||||||
| NC score | -0.002226 (rank : 191) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
SN1L2_MOUSE
|
||||||
| NC score | -0.002522 (rank : 192) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 167 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CFH6 | Gene names | Snf1lk2, Sik2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Salt-inducible kinase 2). | |||||