Please be patient as the page loads
|
PKD2_MOUSE
|
||||||
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PK2L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.967132 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993030 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 7.09276e-130 (rank : 4) | NC score | 0.960207 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 2.78914e-126 (rank : 5) | NC score | 0.958962 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 1.80048e-24 (rank : 6) | NC score | 0.607761 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 2.43343e-21 (rank : 7) | NC score | 0.579567 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 8) | NC score | 0.320461 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 9) | NC score | 0.504520 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
MCLN2_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 10) | NC score | 0.386836 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 11) | NC score | 0.347024 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 12) | NC score | 0.394681 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 13) | NC score | 0.122871 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
MCLN3_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 14) | NC score | 0.380840 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 15) | NC score | 0.345704 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 16) | NC score | 0.348357 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
MCLN3_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 17) | NC score | 0.376855 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 18) | NC score | 0.128288 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
MCLN1_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 19) | NC score | 0.344826 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 20) | NC score | 0.344946 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
MCLN1_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 21) | NC score | 0.349646 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 22) | NC score | 0.341726 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 23) | NC score | 0.342962 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 24) | NC score | 0.296645 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 25) | NC score | 0.332206 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 26) | NC score | 0.300507 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 27) | NC score | 0.207124 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 28) | NC score | 0.341152 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
TRPC3_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 29) | NC score | 0.205584 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 30) | NC score | 0.326012 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 31) | NC score | 0.333459 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 32) | NC score | 0.328247 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 33) | NC score | 0.315342 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 34) | NC score | 0.317749 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 35) | NC score | 0.273798 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 36) | NC score | 0.194228 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 37) | NC score | 0.193814 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 38) | NC score | 0.186230 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 39) | NC score | 0.187014 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 40) | NC score | 0.272170 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 41) | NC score | 0.133161 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 42) | NC score | 0.093618 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.289000 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
TRPC5_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 44) | NC score | 0.161759 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 45) | NC score | 0.161662 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 46) | NC score | 0.104609 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.257510 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 48) | NC score | 0.256729 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 49) | NC score | 0.263361 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 50) | NC score | 0.081743 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 51) | NC score | 0.265334 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 52) | NC score | 0.264726 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 53) | NC score | 0.120447 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 0.163984 (rank : 54) | NC score | 0.033409 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.120204 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.015222 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.037055 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.263264 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 59) | NC score | 0.267666 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 60) | NC score | 0.170412 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 61) | NC score | 0.170521 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |
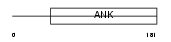
|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 62) | NC score | 0.142585 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.026551 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
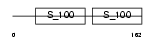
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 64) | NC score | 0.248608 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.025340 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.251857 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.033548 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
NDE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.017337 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.273335 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.263813 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.248246 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
STX4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.014633 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12846, Q15525 | Gene names | STX4, STX4A | |||
|
Domain Architecture |
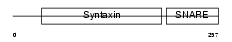
|
|||||
| Description | Syntaxin-4 (NY-REN-31 antigen). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.014765 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.127510 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
CALB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.059728 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |
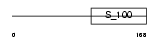
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.009356 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.000053 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
CF152_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.024121 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
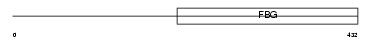
FGL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.007614 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14314 | Gene names | FGL2 | |||
|
Domain Architecture |
|
|||||
| Description | Fibroleukin precursor (Fibrinogen-like protein 2) (pT49). | |||||
|
LSP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.019757 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.007157 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.007351 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
JPH3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.008366 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
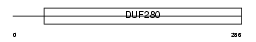
OR5J2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | -0.003076 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 649 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NH18, Q6IEU5 | Gene names | OR5J2 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 5J2 (Olfactory receptor OR11-266). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.013858 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.014150 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
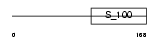
CALB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.053220 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
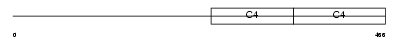
CO4A4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.003964 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |
|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.006104 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.010487 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PDE3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.005839 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
CNTN5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.001047 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P68500 | Gene names | Cntn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.040446 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.008208 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
CAR10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.007017 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
MRGRE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.003185 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZB7, Q711I9 | Gene names | Mrgpre, Ebrt3, Mrge | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mas-related G-protein coupled receptor member E (Evolutionary breakpoint transcript 3 protein). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.005883 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PSMD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.007900 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | -0.002898 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.122003 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.122077 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.118911 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.068312 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.070767 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.068638 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.070124 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.067704 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.064511 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.053742 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.052681 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPV6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.091230 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.086081 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.993030 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.967132 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.960207 (rank : 4) | θ value | 7.09276e-130 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.958962 (rank : 5) | θ value | 2.78914e-126 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.607761 (rank : 6) | θ value | 1.80048e-24 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.579567 (rank : 7) | θ value | 2.43343e-21 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.504520 (rank : 8) | θ value | 3.63628e-17 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.394681 (rank : 9) | θ value | 5.26297e-08 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN2_HUMAN
|
||||||
| NC score | 0.386836 (rank : 10) | θ value | 2.79066e-09 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN3_HUMAN
|
||||||
| NC score | 0.380840 (rank : 11) | θ value | 8.97725e-08 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN3_MOUSE
|
||||||
| NC score | 0.376855 (rank : 12) | θ value | 5.81887e-07 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN1_HUMAN
|
||||||
| NC score | 0.349646 (rank : 13) | θ value | 9.92553e-07 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.348357 (rank : 14) | θ value | 5.81887e-07 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.347024 (rank : 15) | θ value | 3.08544e-08 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.345704 (rank : 16) | θ value | 1.17247e-07 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.344946 (rank : 17) | θ value | 9.92553e-07 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
MCLN1_MOUSE
|
||||||
| NC score | 0.344826 (rank : 18) | θ value | 7.59969e-07 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.342962 (rank : 19) | θ value | 6.43352e-06 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.341726 (rank : 20) | θ value | 2.88788e-06 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.341152 (rank : 21) | θ value | 2.44474e-05 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.333459 (rank : 22) | θ value | 9.29e-05 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.332206 (rank : 23) | θ value | 1.09739e-05 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.328247 (rank : 24) | θ value | 0.000121331 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.326012 (rank : 25) | θ value | 5.44631e-05 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.320461 (rank : 26) | θ value | 1.24977e-17 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.317749 (rank : 27) | θ value | 0.00035302 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.315342 (rank : 28) | θ value | 0.000270298 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.300507 (rank : 29) | θ value | 1.43324e-05 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.296645 (rank : 30) | θ value | 6.43352e-06 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.289000 (rank : 31) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.273798 (rank : 32) | θ value | 0.000602161 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.273335 (rank : 33) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.272170 (rank : 34) | θ value | 0.00298849 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.267666 (rank : 35) | θ value | 0.21417 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.265334 (rank : 36) | θ value | 0.125558 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.264726 (rank : 37) | θ value | 0.125558 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.263813 (rank : 38) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.263361 (rank : 39) | θ value | 0.0961366 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.263264 (rank : 40) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.257510 (rank : 41) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.256729 (rank : 42) | θ value | 0.0563607 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.251857 (rank : 43) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.248608 (rank : 44) | θ value | 0.47712 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.248246 (rank : 45) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.207124 (rank : 46) | θ value | 1.87187e-05 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC3_HUMAN
|
||||||
| NC score | 0.205584 (rank : 47) | θ value | 4.1701e-05 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.194228 (rank : 48) | θ value | 0.00134147 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.193814 (rank : 49) | θ value | 0.00134147 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.187014 (rank : 50) | θ value | 0.00228821 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.186230 (rank : 51) | θ value | 0.00228821 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_MOUSE
|
||||||
| NC score | 0.170521 (rank : 52) | θ value | 0.279714 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.170412 (rank : 53) | θ value | 0.279714 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC5_HUMAN
|
||||||
| NC score | 0.161759 (rank : 54) | θ value | 0.0252991 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.161662 (rank : 55) | θ value | 0.0252991 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.142585 (rank : 56) | θ value | 0.365318 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.133161 (rank : 57) | θ value | 0.00509761 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.128288 (rank : 58) | θ value | 5.81887e-07 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.127510 (rank : 59) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.122871 (rank : 60) | θ value | 5.26297e-08 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.122077 (rank : 61) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.122003 (rank : 62) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.120447 (rank : 63) | θ value | 0.125558 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.120204 (rank : 64) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.118911 (rank : 65) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.104609 (rank : 66) | θ value | 0.0431538 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.093618 (rank : 67) | θ value | 0.0113563 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
TRPV6_HUMAN
|
||||||
| NC score | 0.091230 (rank : 68) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV6_MOUSE
|
||||||
| NC score | 0.086081 (rank : 69) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.081743 (rank : 70) | θ value | 0.0961366 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.070767 (rank : 71) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.070124 (rank : 72) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.068638 (rank : 73) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.068312 (rank : 74) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.067704 (rank : 75) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.064511 (rank : 76) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
CALB1_HUMAN
|
||||||
| NC score | 0.059728 (rank : 77) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
TRPV2_HUMAN
|
||||||
| NC score | 0.053742 (rank : 78) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
CALB1_MOUSE
|
||||||
| NC score | 0.053220 (rank : 79) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.052681 (rank : 80) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.040446 (rank : 81) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.037055 (rank : 82) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.033548 (rank : 83) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.033409 (rank : 84) | θ value | 0.163984 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.026551 (rank : 85) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.025340 (rank : 86) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.024121 (rank : 87) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
LSP1_HUMAN
|
||||||
| NC score | 0.019757 (rank : 88) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
NDE1_HUMAN
|
||||||
| NC score | 0.017337 (rank : 89) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 823 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NXR1, Q49AQ2 | Gene names | NDE1, NUDE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.015222 (rank : 90) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.014765 (rank : 91) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
STX4_HUMAN
|
||||||
| NC score | 0.014633 (rank : 92) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12846, Q15525 | Gene names | STX4, STX4A | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-4 (NY-REN-31 antigen). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.014150 (rank : 93) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.013858 (rank : 94) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.010487 (rank : 95) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.009356 (rank : 96) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
JPH3_HUMAN
|
||||||
| NC score | 0.008366 (rank : 97) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WXH2, Q8N471, Q9HDC3, Q9HDC4 | Gene names | JPH3, JP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-3 (Junctophilin type 3) (JP-3). | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.008208 (rank : 98) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
PSMD2_HUMAN
|
||||||
| NC score | 0.007900 (rank : 99) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
FGL2_HUMAN
|
||||||
| NC score | 0.007614 (rank : 100) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14314 | Gene names | FGL2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibroleukin precursor (Fibrinogen-like protein 2) (pT49). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.007351 (rank : 101) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.007157 (rank : 102) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CAR10_MOUSE
|
||||||
| NC score | 0.007017 (rank : 103) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.006104 (rank : 104) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.005883 (rank : 105) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PDE3B_HUMAN
|
||||||
| NC score | 0.005839 (rank : 106) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13370, O00639, Q14408 | Gene names | PDE3B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-inhibited 3',5'-cyclic phosphodiesterase B (EC 3.1.4.17) (Cyclic GMP-inhibited phosphodiesterase B) (CGI-PDE B) (CGIPDE1) (CGIP1). | |||||
|
CO4A4_HUMAN
|
||||||
| NC score | 0.003964 (rank : 107) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
MRGRE_MOUSE
|
||||||
| NC score | 0.003185 (rank : 108) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZB7, Q711I9 | Gene names | Mrgpre, Ebrt3, Mrge | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mas-related G-protein coupled receptor member E (Evolutionary breakpoint transcript 3 protein). | |||||
|
CNTN5_MOUSE
|
||||||
| NC score | 0.001047 (rank : 109) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P68500 | Gene names | Cntn5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-5 precursor (Neural recognition molecule NB-2). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.000053 (rank : 110) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | -0.002898 (rank : 111) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
OR5J2_HUMAN
|
||||||
| NC score | -0.003076 (rank : 112) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 649 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NH18, Q6IEU5 | Gene names | OR5J2 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 5J2 (Olfactory receptor OR11-266). | |||||