Please be patient as the page loads
|
SCN5A_HUMAN
|
||||||
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SC10A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992667 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993165 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.986495 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.988058 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.986532 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.985924 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.988193 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.987745 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.978532 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.986249 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.985600 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.985736 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 3.70236e-70 (rank : 14) | NC score | 0.862568 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 1.83746e-69 (rank : 15) | NC score | 0.857788 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 1.19101e-68 (rank : 16) | NC score | 0.855339 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 4.52582e-68 (rank : 17) | NC score | 0.858206 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 5.91091e-68 (rank : 18) | NC score | 0.901370 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 1.00825e-67 (rank : 19) | NC score | 0.853561 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 2.93355e-67 (rank : 20) | NC score | 0.857126 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 7.2256e-66 (rank : 21) | NC score | 0.866894 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 1.2325e-65 (rank : 22) | NC score | 0.867231 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 2.74572e-65 (rank : 23) | NC score | 0.850197 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 6.11685e-65 (rank : 24) | NC score | 0.853498 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 3.96481e-64 (rank : 25) | NC score | 0.880581 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 26) | NC score | 0.873663 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 6.76295e-64 (rank : 27) | NC score | 0.852519 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 1.96772e-63 (rank : 28) | NC score | 0.858302 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 5.7252e-63 (rank : 29) | NC score | 0.856883 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 30) | NC score | 0.845448 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 31) | NC score | 0.296645 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 32) | NC score | 0.213394 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 33) | NC score | 0.208099 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 34) | NC score | 0.181178 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 35) | NC score | 0.290354 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 36) | NC score | 0.309196 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 37) | NC score | 0.233566 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 38) | NC score | 0.330244 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 39) | NC score | 0.148035 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 40) | NC score | 0.215547 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 41) | NC score | 0.120938 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 42) | NC score | 0.218912 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 43) | NC score | 0.145661 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.025593 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 45) | NC score | 0.195660 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 46) | NC score | 0.195633 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 47) | NC score | 0.007173 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 48) | NC score | 0.146784 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 49) | NC score | 0.144504 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 50) | NC score | 0.175794 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 51) | NC score | 0.175931 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 52) | NC score | 0.024922 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 53) | NC score | 0.070624 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 54) | NC score | 0.070735 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 55) | NC score | 0.168692 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 56) | NC score | 0.168869 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
CAD16_HUMAN
|
||||||
| θ value | 0.125558 (rank : 57) | NC score | 0.005179 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75309, Q6UW93 | Gene names | CDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 58) | NC score | 0.293199 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 59) | NC score | 0.100556 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 60) | NC score | 0.071343 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
AKAP8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 61) | NC score | 0.016414 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 62) | NC score | 0.121534 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 63) | NC score | 0.119350 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 64) | NC score | 0.128361 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.121539 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 66) | NC score | 0.016044 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 67) | NC score | 0.069531 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 68) | NC score | 0.054684 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
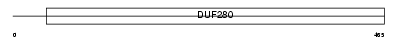
TFE2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 69) | NC score | 0.011662 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.140357 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.022645 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.072703 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 73) | NC score | 0.024247 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
DREB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.012125 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.006437 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.011959 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.020448 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.150476 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
AMPH_MOUSE
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.011767 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 80) | NC score | 0.148746 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.008506 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
OX2R_HUMAN
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | -0.000861 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43614 | Gene names | HCRTR2 | |||
|
Domain Architecture |
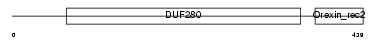
|
|||||
| Description | Orexin receptor type 2 (Ox2r) (Hypocretin receptor type 2). | |||||
|
OX2R_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | -0.000870 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58308, Q5SGC8, Q6VLX3, Q8BG12 | Gene names | Hcrtr2, Mox2r | |||
|
Domain Architecture |
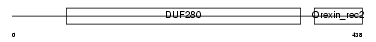
|
|||||
| Description | Orexin receptor type 2 (Ox2r) (Hypocretin receptor type 2). | |||||
|
DNMT1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.013509 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
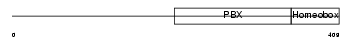
HRH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | -0.000102 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35367 | Gene names | HRH1 | |||
|
Domain Architecture |
|
|||||
| Description | Histamine H1 receptor. | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.017162 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.005056 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.007688 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.027977 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LYRIC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.010768 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
NRIP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.009876 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.006160 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.005388 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
FBX7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.013246 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.127322 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
NCBP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.009328 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q09161, Q59G76, Q5T1V0 | Gene names | NCBP1, CBP80, NCBP | |||
|
Domain Architecture |
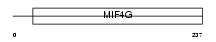
|
|||||
| Description | Nuclear cap-binding protein subunit 1 (80 kDa nuclear cap-binding protein) (NCBP 80 kDa subunit) (CBP80). | |||||
|
NELF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.017264 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6X4W1, Q6X4V7, Q6X4V8, Q6X4V9, Q8N2M2, Q96SY1, Q9NPM4, Q9NPP3, Q9NPS3 | Gene names | NELF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor). | |||||
|
NELF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.019234 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99NF2, Q8BPT0, Q8C9R5, Q9DBF4, Q9ERZ1 | Gene names | Nelf, Jac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor) (Juxtasynaptic attractor of caldendrin on dendritic boutons protein) (Jacob protein). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.017938 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TCO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.010437 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20061, Q8WV77 | Gene names | TCN1, TC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcobalamin-1 precursor (Transcobalamin I) (TCI) (TC I). | |||||
|
GLGB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.008566 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D6Y9 | Gene names | Gbe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.006461 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | -0.000688 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | -0.004837 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.001710 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
RN168_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.003829 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
TPTE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.095420 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.003026 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ADA11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.000349 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |
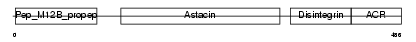
|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.037672 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
ESR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.000400 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03372, Q13511, Q14276, Q9NU51, Q9UDZ7, Q9UIS7 | Gene names | ESR1, ESR, NR3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
OLF9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | -0.001713 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60885, Q8VGC0 | Gene names | Olfr9, Mor269-3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 9 (Olfactory receptor 269-3) (Odorant receptor M25). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.004892 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.002548 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.045170 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.017372 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
ABCA9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.000592 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUA7, Q6P655, Q8N2S4, Q8WWZ5, Q96MD8 | Gene names | ABCA9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 9. | |||||
|
CILP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.001982 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IUL8, Q6NV88, Q8N4A6, Q8WV21 | Gene names | CILP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 2 precursor (CILP-2) [Contains: Cartilage intermediate layer protein 2 C1; Cartilage intermediate layer protein 2 C2]. | |||||
|
CUBN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | -0.001782 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
DLX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | -0.000716 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56177, Q53ZU4, Q8IYB2 | Gene names | DLX1 | |||
|
Domain Architecture |
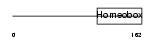
|
|||||
| Description | Homeobox protein DLX-1. | |||||
|
DLX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | -0.000717 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64317 | Gene names | Dlx1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-1. | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.003145 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
TB182_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.009775 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.044232 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.046264 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.035349 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
ARHG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.001161 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
CI061_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.006201 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q4FZH1 | Gene names | Gm967 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf61 homolog. | |||||
|
CSTFT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.006057 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
EPHB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | -0.005286 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54754, Q62214 | Gene names | Ephb3, Etk2, Mdk5, Sek4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-5) (Developmental kinase 5) (SEK-4). | |||||
|
FA26B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.003878 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEC4, Q3TDQ7, Q8BT85, Q8C246 | Gene names | Fam26b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM26B. | |||||
|
FANCG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.005411 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQR6 | Gene names | Fancg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein homolog (Protein FACG). | |||||
|
FCRLA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.002663 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920A9, Q3T9Q2, Q8K3U9, Q8VHP5 | Gene names | Fcrla, Fcrl, Fcrl1, Fcrlm1, Fcrx, Freb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
HXC10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | -0.001444 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
LBXCO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.003419 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
|
MPP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.001183 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
MPP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.001196 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
PHOS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.004192 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20941, Q14816 | Gene names | PDC | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (MEKA protein). | |||||
|
PIGN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.004571 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.000963 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
SC6A5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.000398 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y345, O95288, Q9BX77 | Gene names | SLC6A5, GLYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
SYN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.006572 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZP2 | Gene names | Syn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.011986 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TBX19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.001041 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60806 | Gene names | TBX19, TPIT | |||
|
Domain Architecture |
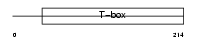
|
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.000317 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.080665 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.082063 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.078650 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.079498 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.071511 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.070537 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.057410 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.057770 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.067777 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.072918 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.073933 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.076684 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.071880 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.065711 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.094516 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.094600 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.064521 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.059465 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.067488 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.077262 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.055948 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.993165 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.992667 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.988193 (rank : 4) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.988058 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.987745 (rank : 6) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.986532 (rank : 7) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.986495 (rank : 8) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.986249 (rank : 9) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.985924 (rank : 10) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.985736 (rank : 11) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.985600 (rank : 12) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.978532 (rank : 13) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.901370 (rank : 14) | θ value | 5.91091e-68 (rank : 18) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.880581 (rank : 15) | θ value | 3.96481e-64 (rank : 25) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.873663 (rank : 16) | θ value | 5.17823e-64 (rank : 26) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.867231 (rank : 17) | θ value | 1.2325e-65 (rank : 22) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.866894 (rank : 18) | θ value | 7.2256e-66 (rank : 21) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.862568 (rank : 19) | θ value | 3.70236e-70 (rank : 14) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.858302 (rank : 20) | θ value | 1.96772e-63 (rank : 28) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.858206 (rank : 21) | θ value | 4.52582e-68 (rank : 17) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.857788 (rank : 22) | θ value | 1.83746e-69 (rank : 15) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.857126 (rank : 23) | θ value | 2.93355e-67 (rank : 20) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.856883 (rank : 24) | θ value | 5.7252e-63 (rank : 29) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.855339 (rank : 25) | θ value | 1.19101e-68 (rank : 16) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.853561 (rank : 26) | θ value | 1.00825e-67 (rank : 19) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.853498 (rank : 27) | θ value | 6.11685e-65 (rank : 24) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.852519 (rank : 28) | θ value | 6.76295e-64 (rank : 27) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.850197 (rank : 29) | θ value | 2.74572e-65 (rank : 23) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.845448 (rank : 30) | θ value | 2.48917e-58 (rank : 30) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.330244 (rank : 31) | θ value | 0.000461057 (rank : 38) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.309196 (rank : 32) | θ value | 0.00035302 (rank : 36) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.296645 (rank : 33) | θ value | 6.43352e-06 (rank : 31) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.293199 (rank : 34) | θ value | 0.125558 (rank : 58) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.290354 (rank : 35) | θ value | 9.29e-05 (rank : 35) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.233566 (rank : 36) | θ value | 0.000461057 (rank : 37) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.218912 (rank : 37) | θ value | 0.00298849 (rank : 42) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.215547 (rank : 38) | θ value | 0.00102713 (rank : 40) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.213394 (rank : 39) | θ value | 2.44474e-05 (rank : 32) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.208099 (rank : 40) | θ value | 5.44631e-05 (rank : 33) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.195660 (rank : 41) | θ value | 0.0193708 (rank : 45) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.195633 (rank : 42) | θ value | 0.0193708 (rank : 46) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.181178 (rank : 43) | θ value | 5.44631e-05 (rank : 34) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.175931 (rank : 44) | θ value | 0.0563607 (rank : 51) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.175794 (rank : 45) | θ value | 0.0563607 (rank : 50) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.168869 (rank : 46) | θ value | 0.0736092 (rank : 56) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.168692 (rank : 47) | θ value | 0.0736092 (rank : 55) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.150476 (rank : 48) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.148746 (rank : 49) | θ value | 1.06291 (rank : 80) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.148035 (rank : 50) | θ value | 0.000602161 (rank : 39) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.146784 (rank : 51) | θ value | 0.0431538 (rank : 48) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.145661 (rank : 52) | θ value | 0.0113563 (rank : 43) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.144504 (rank : 53) | θ value | 0.0563607 (rank : 49) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.140357 (rank : 54) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.128361 (rank : 55) | θ value | 0.279714 (rank : 64) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.127322 (rank : 56) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.121539 (rank : 57) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.121534 (rank : 58) | θ value | 0.21417 (rank : 62) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.120938 (rank : 59) | θ value | 0.00175202 (rank : 41) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.119350 (rank : 60) | θ value | 0.279714 (rank : 63) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.100556 (rank : 61) | θ value | 0.125558 (rank : 59) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.095420 (rank : 62) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.094600 (rank : 63) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.094516 (rank : 64) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.082063 (rank : 65) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.080665 (rank : 66) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.079498 (rank : 67) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.078650 (rank : 68) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.077262 (rank : 69) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.076684 (rank : 70) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.073933 (rank : 71) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.072918 (rank : 72) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.072703 (rank : 73) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.071880 (rank : 74) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.071511 (rank : 75) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.071343 (rank : 76) | θ value | 0.163984 (rank : 60) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.070735 (rank : 77) | θ value | 0.0563607 (rank : 54) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.070624 (rank : 78) | θ value | 0.0563607 (rank : 53) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.070537 (rank : 79) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.069531 (rank : 80) | θ value | 0.279714 (rank : 67) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.067777 (rank : 81) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.067488 (rank : 82) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.065711 (rank : 83) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.064521 (rank : 84) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.059465 (rank : 85) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.057770 (rank : 86) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.057410 (rank : 87) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.055948 (rank : 88) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.054684 (rank : 89) | θ value | 0.365318 (rank : 68) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.046264 (rank : 90) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.045170 (rank : 91) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.044232 (rank : 92) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.037672 (rank : 93) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.035349 (rank : 94) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.027977 (rank : 95) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.025593 (rank : 96) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.024922 (rank : 97) | θ value | 0.0563607 (rank : 52) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.024247 (rank : 98) | θ value | 0.62314 (rank : 73) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.022645 (rank : 99) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.020448 (rank : 100) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NELF_MOUSE
|
||||||
| NC score | 0.019234 (rank : 101) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99NF2, Q8BPT0, Q8C9R5, Q9DBF4, Q9ERZ1 | Gene names | Nelf, Jac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor) (Juxtasynaptic attractor of caldendrin on dendritic boutons protein) (Jacob protein). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.017938 (rank : 102) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.017372 (rank : 103) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
NELF_HUMAN
|
||||||
| NC score | 0.017264 (rank : 104) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6X4W1, Q6X4V7, Q6X4V8, Q6X4V9, Q8N2M2, Q96SY1, Q9NPM4, Q9NPP3, Q9NPS3 | Gene names | NELF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nasal embryonic luteinizing hormone-releasing hormone factor (Nasal embryonic LHRH factor). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.017162 (rank : 105) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
AKAP8_HUMAN
|
||||||
| NC score | 0.016414 (rank : 106) | θ value | 0.21417 (rank : 61) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43823 | Gene names | AKAP8, AKAP95 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.016044 (rank : 107) | θ value | 0.279714 (rank : 66) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
DNMT1_MOUSE
|
||||||
| NC score | 0.013509 (rank : 108) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
FBX7_HUMAN
|
||||||
| NC score | 0.013246 (rank : 109) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
DREB_HUMAN
|
||||||
| NC score | 0.012125 (rank : 110) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16643 | Gene names | DBN1 | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.011986 (rank : 111) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.011959 (rank : 112) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
AMPH_MOUSE
|
||||||
| NC score | 0.011767 (rank : 113) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQF7, Q8R1C4 | Gene names | Amph, Amph1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
TFE2_HUMAN
|
||||||
| NC score | 0.011662 (rank : 114) | θ value | 0.365318 (rank : 69) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
LYRIC_MOUSE
|
||||||
| NC score | 0.010768 (rank : 115) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80WJ7, Q3U9F8, Q3UAQ8, Q8BN67, Q8CBT9, Q8CDL0, Q8CGI7, Q9D052 | Gene names | Mtdh, Lyric | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/LYRIC) (Metastasis adhesion protein) (Metadherin). | |||||
|
TCO1_HUMAN
|
||||||
| NC score | 0.010437 (rank : 116) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20061, Q8WV77 | Gene names | TCN1, TC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcobalamin-1 precursor (Transcobalamin I) (TCI) (TC I). | |||||
|
NRIP1_MOUSE
|
||||||
| NC score | 0.009876 (rank : 117) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.009775 (rank : 118) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
NCBP1_HUMAN
|
||||||
| NC score | 0.009328 (rank : 119) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q09161, Q59G76, Q5T1V0 | Gene names | NCBP1, CBP80, NCBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear cap-binding protein subunit 1 (80 kDa nuclear cap-binding protein) (NCBP 80 kDa subunit) (CBP80). | |||||
|
GLGB_MOUSE
|
||||||
| NC score | 0.008566 (rank : 120) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D6Y9 | Gene names | Gbe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.008506 (rank : 121) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.007688 (rank : 122) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.007173 (rank : 123) | θ value | 0.0252991 (rank : 47) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
SYN3_MOUSE
|
||||||
| NC score | 0.006572 (rank : 124) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8JZP2 | Gene names | Syn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-3 (Synapsin III). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.006461 (rank : 125) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.006437 (rank : 126) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
CI061_MOUSE
|
||||||
| NC score | 0.006201 (rank : 127) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q4FZH1 | Gene names | Gm967 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf61 homolog. | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.006160 (rank : 128) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
CSTFT_MOUSE
|
||||||
| NC score | 0.006057 (rank : 129) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C7E9, Q6A016, Q8BHH7, Q8C3W7, Q8R2Y1, Q9EPU3 | Gene names | Cstf2t, Kiaa0689 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage stimulation factor 64 kDa subunit, tau variant (CSTF 64 kDa subunit, tau variant) (CF-1 64 kDa subunit, tau variant) (TauCstF-64). | |||||
|
FANCG_MOUSE
|
||||||
| NC score | 0.005411 (rank : 130) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQR6 | Gene names | Fancg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group G protein homolog (Protein FACG). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.005388 (rank : 131) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CAD16_HUMAN
|
||||||
| NC score | 0.005179 (rank : 132) | θ value | 0.125558 (rank : 57) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75309, Q6UW93 | Gene names | CDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.005056 (rank : 133) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.004892 (rank : 134) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
PIGN_MOUSE
|
||||||
| NC score | 0.004571 (rank : 135) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
PHOS_HUMAN
|
||||||
| NC score | 0.004192 (rank : 136) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20941, Q14816 | Gene names | PDC | |||
|
Domain Architecture |

|
|||||
| Description | Phosducin (PHD) (33 kDa phototransducing protein) (MEKA protein). | |||||
|
FA26B_MOUSE
|
||||||
| NC score | 0.003878 (rank : 137) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VEC4, Q3TDQ7, Q8BT85, Q8C246 | Gene names | Fam26b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM26B. | |||||
|
RN168_HUMAN
|
||||||
| NC score | 0.003829 (rank : 138) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
LBXCO_MOUSE
|
||||||
| NC score | 0.003419 (rank : 139) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BX46, Q3UYA4, Q5W8I2, Q8C0T2 | Gene names | Lbxcor1, Corl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladybird homeobox corepressor 1 (Transcriptional corepressor Corl1). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.003145 (rank : 140) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.003026 (rank : 141) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
FCRLA_MOUSE
|
||||||
| NC score | 0.002663 (rank : 142) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920A9, Q3T9Q2, Q8K3U9, Q8VHP5 | Gene names | Fcrla, Fcrl, Fcrl1, Fcrlm1, Fcrx, Freb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fc receptor-like and mucin-like 1 precursor (Fc receptor-like A) (Fc receptor homolog expressed in B-cells) (Fc receptor-related protein X) (FcRX) (Fc receptor-like protein). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.002548 (rank : 143) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
CILP2_HUMAN
|
||||||
| NC score | 0.001982 (rank : 144) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IUL8, Q6NV88, Q8N4A6, Q8WV21 | Gene names | CILP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cartilage intermediate layer protein 2 precursor (CILP-2) [Contains: Cartilage intermediate layer protein 2 C1; Cartilage intermediate layer protein 2 C2]. | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.001710 (rank : 145) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
MPP5_MOUSE
|
||||||
| NC score | 0.001196 (rank : 146) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
MPP5_HUMAN
|
||||||
| NC score | 0.001183 (rank : 147) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
ARHG1_HUMAN
|
||||||
| NC score | 0.001161 (rank : 148) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
TBX19_HUMAN
|
||||||
| NC score | 0.001041 (rank : 149) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60806 | Gene names | TBX19, TPIT | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.000963 (rank : 150) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
ABCA9_HUMAN
|
||||||
| NC score | 0.000592 (rank : 151) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUA7, Q6P655, Q8N2S4, Q8WWZ5, Q96MD8 | Gene names | ABCA9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 9. | |||||
|
ESR1_HUMAN
|
||||||
| NC score | 0.000400 (rank : 152) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03372, Q13511, Q14276, Q9NU51, Q9UDZ7, Q9UIS7 | Gene names | ESR1, ESR, NR3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
SC6A5_HUMAN
|
||||||
| NC score | 0.000398 (rank : 153) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y345, O95288, Q9BX77 | Gene names | SLC6A5, GLYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
ADA11_HUMAN
|
||||||
| NC score | 0.000349 (rank : 154) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.000317 (rank : 155) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
HRH1_HUMAN
|
||||||
| NC score | -0.000102 (rank : 156) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35367 | Gene names | HRH1 | |||
|
Domain Architecture |

|
|||||
| Description | Histamine H1 receptor. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | -0.000688 (rank : 157) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
DLX1_HUMAN
|
||||||
| NC score | -0.000716 (rank : 158) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56177, Q53ZU4, Q8IYB2 | Gene names | DLX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-1. | |||||
|
DLX1_MOUSE
|
||||||
| NC score | -0.000717 (rank : 159) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64317 | Gene names | Dlx1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-1. | |||||
|
OX2R_HUMAN
|
||||||
| NC score | -0.000861 (rank : 160) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43614 | Gene names | HCRTR2 | |||
|
Domain Architecture |

|
|||||
| Description | Orexin receptor type 2 (Ox2r) (Hypocretin receptor type 2). | |||||
|
OX2R_MOUSE
|
||||||
| NC score | -0.000870 (rank : 161) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58308, Q5SGC8, Q6VLX3, Q8BG12 | Gene names | Hcrtr2, Mox2r | |||
|
Domain Architecture |

|
|||||
| Description | Orexin receptor type 2 (Ox2r) (Hypocretin receptor type 2). | |||||
|
HXC10_HUMAN
|
||||||
| NC score | -0.001444 (rank : 162) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYD6, O15219, O15220, Q9BVD5 | Gene names | HOXC10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C10. | |||||
|
OLF9_MOUSE
|
||||||
| NC score | -0.001713 (rank : 163) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60885, Q8VGC0 | Gene names | Olfr9, Mor269-3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 9 (Olfactory receptor 269-3) (Odorant receptor M25). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | -0.001782 (rank : 164) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | -0.004837 (rank : 165) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
EPHB3_MOUSE
|
||||||
| NC score | -0.005286 (rank : 166) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 145 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54754, Q62214 | Gene names | Ephb3, Etk2, Mdk5, Sek4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-5) (Developmental kinase 5) (SEK-4). | |||||