Please be patient as the page loads
|
SCN9A_HUMAN
|
||||||
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
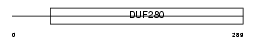
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SC10A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991707 (rank : 8) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991771 (rank : 7) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.982985 (rank : 13) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.984696 (rank : 12) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994925 (rank : 4) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.995157 (rank : 3) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.995789 (rank : 2) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.990950 (rank : 9) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.985736 (rank : 10) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.985220 (rank : 11) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.993884 (rank : 6) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.994332 (rank : 5) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 157 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 1.31376e-75 (rank : 14) | NC score | 0.893607 (rank : 14) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 1.60597e-73 (rank : 15) | NC score | 0.851713 (rank : 19) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 1.35953e-72 (rank : 16) | NC score | 0.845888 (rank : 22) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 5.1662e-72 (rank : 17) | NC score | 0.843979 (rank : 25) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 6.7473e-72 (rank : 18) | NC score | 0.872989 (rank : 15) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 8.81223e-72 (rank : 19) | NC score | 0.846393 (rank : 21) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 1.96316e-71 (rank : 20) | NC score | 0.842207 (rank : 26) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 1.66191e-70 (rank : 21) | NC score | 0.866034 (rank : 16) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 2.83479e-70 (rank : 22) | NC score | 0.855635 (rank : 18) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 3.70236e-70 (rank : 23) | NC score | 0.841119 (rank : 27) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 4.83544e-70 (rank : 24) | NC score | 0.856030 (rank : 17) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 4.83544e-70 (rank : 25) | NC score | 0.840279 (rank : 28) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 3.13423e-69 (rank : 26) | NC score | 0.845106 (rank : 23) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 3.4653e-68 (rank : 27) | NC score | 0.838679 (rank : 29) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 1.00825e-67 (rank : 28) | NC score | 0.848899 (rank : 20) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 3.83135e-67 (rank : 29) | NC score | 0.845017 (rank : 24) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 1.04338e-64 (rank : 30) | NC score | 0.833834 (rank : 30) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 31) | NC score | 0.216165 (rank : 37) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 32) | NC score | 0.196508 (rank : 42) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 33) | NC score | 0.196544 (rank : 41) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 34) | NC score | 0.309094 (rank : 31) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 35) | NC score | 0.274068 (rank : 33) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 36) | NC score | 0.231756 (rank : 36) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 37) | NC score | 0.207499 (rank : 39) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 38) | NC score | 0.202245 (rank : 40) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 39) | NC score | 0.215412 (rank : 38) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 40) | NC score | 0.170337 (rank : 47) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 41) | NC score | 0.170500 (rank : 46) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 42) | NC score | 0.018691 (rank : 101) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 43) | NC score | 0.264411 (rank : 35) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 44) | NC score | 0.018899 (rank : 100) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 45) | NC score | 0.177769 (rank : 44) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 46) | NC score | 0.177835 (rank : 43) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 47) | NC score | 0.175777 (rank : 45) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 48) | NC score | 0.285318 (rank : 32) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 49) | NC score | 0.018975 (rank : 99) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 50) | NC score | 0.144416 (rank : 50) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 51) | NC score | 0.012937 (rank : 119) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 52) | NC score | 0.142097 (rank : 51) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 53) | NC score | 0.151678 (rank : 48) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 54) | NC score | 0.149867 (rank : 49) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 55) | NC score | 0.080975 (rank : 65) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 56) | NC score | 0.094871 (rank : 60) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 57) | NC score | 0.017762 (rank : 104) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 58) | NC score | 0.012538 (rank : 123) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 59) | NC score | 0.067434 (rank : 77) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 60) | NC score | 0.067327 (rank : 78) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
MYPT1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 61) | NC score | 0.008853 (rank : 142) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.023443 (rank : 95) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.008915 (rank : 141) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 64) | NC score | 0.017175 (rank : 105) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 65) | NC score | 0.267666 (rank : 34) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 0.365318 (rank : 66) | NC score | 0.018568 (rank : 102) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.138354 (rank : 53) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 68) | NC score | 0.041928 (rank : 90) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.057490 (rank : 88) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 70) | NC score | 0.016179 (rank : 107) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 71) | NC score | 0.014732 (rank : 115) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.118190 (rank : 57) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.093349 (rank : 62) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.015417 (rank : 112) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.116003 (rank : 59) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.139996 (rank : 52) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.019880 (rank : 97) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.015547 (rank : 111) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
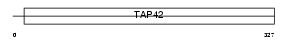
IGBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.014312 (rank : 116) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61249 | Gene names | Igbp1, Pc52 | |||
|
Domain Architecture |
|
|||||
| Description | Immunoglobulin-binding protein 1 (CD79a-binding protein 1) (Alpha4 phosphoprotein) (Lymphocyte signal transduction molecule alpha 4) (p52). | |||||
|
TBX3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.004901 (rank : 154) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 81) | NC score | 0.018132 (rank : 103) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 82) | NC score | 0.016797 (rank : 106) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.004079 (rank : 157) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.002796 (rank : 162) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.003307 (rank : 158) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 86) | NC score | 0.015226 (rank : 113) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 87) | NC score | 0.015987 (rank : 108) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.011044 (rank : 131) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.041473 (rank : 91) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
CASC5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | 0.012644 (rank : 121) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
CPSF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.010774 (rank : 134) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.011152 (rank : 129) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.125661 (rank : 54) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.118765 (rank : 56) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
SON_MOUSE
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.011651 (rank : 127) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TANK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.012735 (rank : 120) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70347, Q61178 | Gene names | Tank, Itraf | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.011911 (rank : 125) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.061296 (rank : 85) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.062808 (rank : 84) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
ANPRA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.002513 (rank : 164) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.002516 (rank : 163) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
CCD93_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.013825 (rank : 118) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
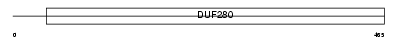
HRH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | -0.001023 (rank : 169) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35367 | Gene names | HRH1 | |||
|
Domain Architecture |
|
|||||
| Description | Histamine H1 receptor. | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.006332 (rank : 149) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.003182 (rank : 160) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.066529 (rank : 80) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
CMTA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.012551 (rank : 122) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.118177 (rank : 58) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
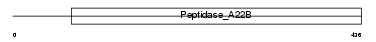
PSN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.033432 (rank : 92) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61144, O54977, P97934, P97935, Q91VS3, Q9D616 | Gene names | Psen2, Ad4h, Alg3, Ps-2, Psnl2 | |||
|
Domain Architecture |
|
|||||
| Description | Presenilin-2 (EC 3.4.23.-) (PS-2) [Contains: Presenilin-2 NTF subunit; Presenilin-2 CTF subunit]. | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.012133 (rank : 124) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
ZRAB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.015554 (rank : 110) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
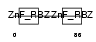
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
AT5F1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.009683 (rank : 138) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24539, Q9BQ68, Q9BRU8 | Gene names | ATP5F1 | |||
|
Domain Architecture |
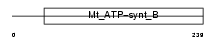
|
|||||
| Description | ATP synthase B chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.011895 (rank : 126) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.007443 (rank : 144) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.026822 (rank : 94) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.011049 (rank : 130) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
TPTE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.094434 (rank : 61) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
ABRA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.007625 (rank : 143) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BUZ1, Q3UU01, Q8BLH3, Q8K431 | Gene names | Abra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.010768 (rank : 135) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ATF6B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.005741 (rank : 152) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.010914 (rank : 132) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.072378 (rank : 72) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.092981 (rank : 63) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.121820 (rank : 55) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
LGR4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | -0.001168 (rank : 170) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXB1, Q9NYD1 | Gene names | LGR4, GPR48 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 4 precursor (G-protein coupled receptor 48). | |||||
|
MEPE_HUMAN
|
||||||
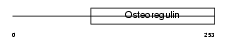
| θ value | 5.27518 (rank : 126) | NC score | 0.028849 (rank : 93) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |
|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.010687 (rank : 136) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
OX2R_HUMAN
|
||||||
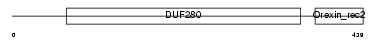
| θ value | 5.27518 (rank : 128) | NC score | -0.002590 (rank : 172) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43614 | Gene names | HCRTR2 | |||
|
Domain Architecture |
|
|||||
| Description | Orexin receptor type 2 (Ox2r) (Hypocretin receptor type 2). | |||||
|
OX2R_MOUSE
|
||||||
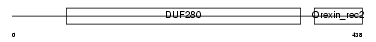
| θ value | 5.27518 (rank : 129) | NC score | -0.002616 (rank : 173) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58308, Q5SGC8, Q6VLX3, Q8BG12 | Gene names | Hcrtr2, Mox2r | |||
|
Domain Architecture |
|
|||||
| Description | Orexin receptor type 2 (Ox2r) (Hypocretin receptor type 2). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.009421 (rank : 139) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TRAK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.010895 (rank : 133) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
AF9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.006853 (rank : 145) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
AT1A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.000521 (rank : 168) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
CR034_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.014099 (rank : 117) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
DIP2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.004228 (rank : 156) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.011253 (rank : 128) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.004270 (rank : 155) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.002366 (rank : 165) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
MARH4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.005919 (rank : 150) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E8, Q86WR8 | Gene names | MARCH4, KIAA1399, RNF174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated RING finger protein 4 (Membrane-associated RING-CH protein IV) (MARCH-IV) (RING finger protein 174). | |||||
|
NGAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.006461 (rank : 147) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
PI2R_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.003070 (rank : 161) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P43119 | Gene names | PTGIR, PRIPR | |||
|
Domain Architecture |
|
|||||
| Description | Prostacyclin receptor (Prostanoid IP receptor) (PGI receptor) (Prostaglandin I2 receptor). | |||||
|
PSD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.005063 (rank : 153) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.015713 (rank : 109) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.019843 (rank : 98) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.020224 (rank : 96) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.006453 (rank : 148) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CY15A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.006645 (rank : 146) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZA5 | Gene names | CYorf15A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative testis protein CYorf15A. | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.009286 (rank : 140) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.003231 (rank : 159) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
K0831_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.005916 (rank : 151) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZNE5, O94920 | Gene names | KIAA0831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
KIF1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.000653 (rank : 167) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
KLF5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | -0.005019 (rank : 174) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13887, Q9UHP8 | Gene names | KLF5, BTEB2, CKLF, IKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Colon krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2) (GC box-binding protein 2). | |||||
|
MPP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.000872 (rank : 166) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.009905 (rank : 137) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
OLF9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | -0.001771 (rank : 171) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60885, Q8VGC0 | Gene names | Olfr9, Mor269-3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 9 (Olfactory receptor 269-3) (Odorant receptor M25). | |||||
|
SEGN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.014780 (rank : 114) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |
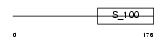
|
|||||
| Description | Secretagogin. | |||||
|
ZN184_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | -0.005540 (rank : 175) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99676, O60792, Q8TBA9 | Gene names | ZNF184 | |||
|
Domain Architecture |
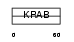
|
|||||
| Description | Zinc finger protein 184. | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.079598 (rank : 67) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.080950 (rank : 66) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.077560 (rank : 69) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.078390 (rank : 68) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.070679 (rank : 75) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.069827 (rank : 76) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.057425 (rank : 89) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.057840 (rank : 87) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.067158 (rank : 79) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.072057 (rank : 73) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.072865 (rank : 71) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.075654 (rank : 70) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.065761 (rank : 81) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.092539 (rank : 64) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.064120 (rank : 82) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.063414 (rank : 83) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.071643 (rank : 74) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.058019 (rank : 86) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 157 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.995789 (rank : 2) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.995157 (rank : 3) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.994925 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.994332 (rank : 5) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.993884 (rank : 6) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.991771 (rank : 7) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.991707 (rank : 8) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.990950 (rank : 9) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.985736 (rank : 10) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.985220 (rank : 11) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.984696 (rank : 12) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.982985 (rank : 13) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.893607 (rank : 14) | θ value | 1.31376e-75 (rank : 14) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.872989 (rank : 15) | θ value | 6.7473e-72 (rank : 18) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.866034 (rank : 16) | θ value | 1.66191e-70 (rank : 21) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.856030 (rank : 17) | θ value | 4.83544e-70 (rank : 24) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.855635 (rank : 18) | θ value | 2.83479e-70 (rank : 22) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.851713 (rank : 19) | θ value | 1.60597e-73 (rank : 15) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.848899 (rank : 20) | θ value | 1.00825e-67 (rank : 28) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.846393 (rank : 21) | θ value | 8.81223e-72 (rank : 19) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.845888 (rank : 22) | θ value | 1.35953e-72 (rank : 16) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.845106 (rank : 23) | θ value | 3.13423e-69 (rank : 26) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.845017 (rank : 24) | θ value | 3.83135e-67 (rank : 29) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.843979 (rank : 25) | θ value | 5.1662e-72 (rank : 17) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.842207 (rank : 26) | θ value | 1.96316e-71 (rank : 20) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.841119 (rank : 27) | θ value | 3.70236e-70 (rank : 23) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.840279 (rank : 28) | θ value | 4.83544e-70 (rank : 25) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.838679 (rank : 29) | θ value | 3.4653e-68 (rank : 27) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.833834 (rank : 30) | θ value | 1.04338e-64 (rank : 30) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.309094 (rank : 31) | θ value | 0.000158464 (rank : 34) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.285318 (rank : 32) | θ value | 0.0113563 (rank : 48) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.274068 (rank : 33) | θ value | 0.000602161 (rank : 35) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.267666 (rank : 34) | θ value | 0.21417 (rank : 65) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.264411 (rank : 35) | θ value | 0.00869519 (rank : 43) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.231756 (rank : 36) | θ value | 0.00175202 (rank : 36) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.216165 (rank : 37) | θ value | 9.29e-05 (rank : 31) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.215412 (rank : 38) | θ value | 0.00298849 (rank : 39) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.207499 (rank : 39) | θ value | 0.00175202 (rank : 37) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.202245 (rank : 40) | θ value | 0.00298849 (rank : 38) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.196544 (rank : 41) | θ value | 0.000158464 (rank : 33) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.196508 (rank : 42) | θ value | 0.000158464 (rank : 32) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.177835 (rank : 43) | θ value | 0.0113563 (rank : 46) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.177769 (rank : 44) | θ value | 0.0113563 (rank : 45) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.175777 (rank : 45) | θ value | 0.0113563 (rank : 47) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.170500 (rank : 46) | θ value | 0.00509761 (rank : 41) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.170337 (rank : 47) | θ value | 0.00509761 (rank : 40) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.151678 (rank : 48) | θ value | 0.0431538 (rank : 53) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.149867 (rank : 49) | θ value | 0.0563607 (rank : 54) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.144416 (rank : 50) | θ value | 0.0330416 (rank : 50) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.142097 (rank : 51) | θ value | 0.0431538 (rank : 52) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.139996 (rank : 52) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.138354 (rank : 53) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.125661 (rank : 54) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.121820 (rank : 55) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.118765 (rank : 56) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.118190 (rank : 57) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.118177 (rank : 58) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.116003 (rank : 59) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.094871 (rank : 60) | θ value | 0.0961366 (rank : 56) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.094434 (rank : 61) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.093349 (rank : 62) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.092981 (rank : 63) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.092539 (rank : 64) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.080975 (rank : 65) | θ value | 0.0961366 (rank : 55) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.080950 (rank : 66) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.079598 (rank : 67) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.078390 (rank : 68) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.077560 (rank : 69) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.075654 (rank : 70) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.072865 (rank : 71) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.072378 (rank : 72) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.072057 (rank : 73) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.071643 (rank : 74) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.070679 (rank : 75) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.069827 (rank : 76) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.067434 (rank : 77) | θ value | 0.125558 (rank : 59) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.067327 (rank : 78) | θ value | 0.125558 (rank : 60) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.067158 (rank : 79) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.066529 (rank : 80) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.065761 (rank : 81) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.064120 (rank : 82) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.063414 (rank : 83) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.062808 (rank : 84) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.061296 (rank : 85) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.058019 (rank : 86) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.057840 (rank : 87) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.057490 (rank : 88) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.057425 (rank : 89) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.041928 (rank : 90) | θ value | 0.47712 (rank : 68) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.041473 (rank : 91) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
PSN2_MOUSE
|
||||||
| NC score | 0.033432 (rank : 92) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61144, O54977, P97934, P97935, Q91VS3, Q9D616 | Gene names | Psen2, Ad4h, Alg3, Ps-2, Psnl2 | |||
|
Domain Architecture |

|
|||||
| Description | Presenilin-2 (EC 3.4.23.-) (PS-2) [Contains: Presenilin-2 NTF subunit; Presenilin-2 CTF subunit]. | |||||
|
MEPE_HUMAN
|
||||||
| NC score | 0.028849 (rank : 93) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQ76 | Gene names | MEPE | |||
|
Domain Architecture |

|
|||||
| Description | Matrix extracellular phosphoglycoprotein precursor (Osteoblast/osteocyte factor 45) (OF45). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.026822 (rank : 94) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.023443 (rank : 95) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.020224 (rank : 96) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.019880 (rank : 97) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.019843 (rank : 98) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.018975 (rank : 99) | θ value | 0.0113563 (rank : 49) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.018899 (rank : 100) | θ value | 0.0113563 (rank : 44) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.018691 (rank : 101) | θ value | 0.00509761 (rank : 42) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.018568 (rank : 102) | θ value | 0.365318 (rank : 66) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.018132 (rank : 103) | θ value | 0.813845 (rank : 81) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.017762 (rank : 104) | θ value | 0.125558 (rank : 57) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.017175 (rank : 105) | θ value | 0.21417 (rank : 64) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.016797 (rank : 106) | θ value | 0.813845 (rank : 82) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.016179 (rank : 107) | θ value | 0.47712 (rank : 70) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.015987 (rank : 108) | θ value | 1.06291 (rank : 87) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.015713 (rank : 109) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ZRAB2_HUMAN
|
||||||
| NC score | 0.015554 (rank : 110) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
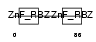
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.015547 (rank : 111) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.015417 (rank : 112) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.015226 (rank : 113) | θ value | 1.06291 (rank : 86) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
SEGN_HUMAN
|
||||||
| NC score | 0.014780 (rank : 114) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.014732 (rank : 115) | θ value | 0.47712 (rank : 71) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
IGBP1_MOUSE
|
||||||
| NC score | 0.014312 (rank : 116) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61249 | Gene names | Igbp1, Pc52 | |||
|
Domain Architecture |

|
|||||
| Description | Immunoglobulin-binding protein 1 (CD79a-binding protein 1) (Alpha4 phosphoprotein) (Lymphocyte signal transduction molecule alpha 4) (p52). | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.014099 (rank : 117) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CCD93_HUMAN
|
||||||
| NC score | 0.013825 (rank : 118) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.012937 (rank : 119) | θ value | 0.0431538 (rank : 51) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TANK_MOUSE
|
||||||
| NC score | 0.012735 (rank : 120) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70347, Q61178 | Gene names | Tank, Itraf | |||
|
Domain Architecture |

|
|||||
| Description | TRAF family member-associated NF-kappa-B activator (TRAF-interacting protein) (I-TRAF). | |||||
|
CASC5_MOUSE
|
||||||
| NC score | 0.012644 (rank : 121) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
CMTA1_HUMAN
|
||||||
| NC score | 0.012551 (rank : 122) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.012538 (rank : 123) | θ value | 0.125558 (rank : 58) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.012133 (rank : 124) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.011911 (rank : 125) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.011895 (rank : 126) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.011651 (rank : 127) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.011253 (rank : 128) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.011152 (rank : 129) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.011049 (rank : 130) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.011044 (rank : 131) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.010914 (rank : 132) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
TRAK1_HUMAN
|
||||||
| NC score | 0.010895 (rank : 133) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPV9, Q96B69 | Gene names | TRAK1, KIAA1042, OIP106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trafficking kinesin-binding protein 1 (106 kDa O-GlcNAc transferase- interacting protein). | |||||
|
CPSF1_HUMAN
|
||||||
| NC score | 0.010774 (rank : 134) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.010768 (rank : 135) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.010687 (rank : 136) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.009905 (rank : 137) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
AT5F1_HUMAN
|
||||||
| NC score | 0.009683 (rank : 138) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24539, Q9BQ68, Q9BRU8 | Gene names | ATP5F1 | |||
|
Domain Architecture |
|
|||||
| Description | ATP synthase B chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.009421 (rank : 139) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.009286 (rank : 140) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.008915 (rank : 141) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
MYPT1_MOUSE
|
||||||
| NC score | 0.008853 (rank : 142) | θ value | 0.163984 (rank : 61) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9DBR7, Q8CBV2, Q99NB6 | Gene names | Ppp1r12a, Mypt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1). | |||||
|
ABRA_MOUSE
|
||||||
| NC score | 0.007625 (rank : 143) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BUZ1, Q3UU01, Q8BLH3, Q8K431 | Gene names | Abra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.007443 (rank : 144) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.006853 (rank : 145) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
CY15A_HUMAN
|
||||||
| NC score | 0.006645 (rank : 146) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZA5 | Gene names | CYorf15A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative testis protein CYorf15A. | |||||
|
NGAP_HUMAN
|
||||||
| NC score | 0.006461 (rank : 147) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UJF2, O95174, Q5TFU9 | Gene names | RASAL2, NGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein nGAP (RAS protein activator-like 1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.006453 (rank : 148) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.006332 (rank : 149) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
MARH4_HUMAN
|
||||||
| NC score | 0.005919 (rank : 150) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2E8, Q86WR8 | Gene names | MARCH4, KIAA1399, RNF174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated RING finger protein 4 (Membrane-associated RING-CH protein IV) (MARCH-IV) (RING finger protein 174). | |||||
|
K0831_HUMAN
|
||||||
| NC score | 0.005916 (rank : 151) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZNE5, O94920 | Gene names | KIAA0831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
ATF6B_MOUSE
|
||||||
| NC score | 0.005741 (rank : 152) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
PSD4_HUMAN
|
||||||
| NC score | 0.005063 (rank : 153) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDX1, O95621, Q4ZG34, Q6GPH8, Q8IYP4 | Gene names | PSD4, EFA6B, TIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH and SEC7 domain-containing protein 4 (Pleckstrin homology and SEC7 domain-containing protein 4) (Exchange factor for ADP-ribosylation factor guanine nucleotide factor 6B) (Telomeric of interleukin-1 cluster protein). | |||||
|
TBX3_HUMAN
|
||||||
| NC score | 0.004901 (rank : 154) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.004270 (rank : 155) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
DIP2A_MOUSE
|
||||||
| NC score | 0.004228 (rank : 156) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWT5, Q8CHI0 | Gene names | Dip2a, Dip2, Kiaa0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.004079 (rank : 157) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.003307 (rank : 158) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.003231 (rank : 159) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.003182 (rank : 160) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
PI2R_HUMAN
|
||||||
| NC score | 0.003070 (rank : 161) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P43119 | Gene names | PTGIR, PRIPR | |||
|
Domain Architecture |

|
|||||
| Description | Prostacyclin receptor (Prostanoid IP receptor) (PGI receptor) (Prostaglandin I2 receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.002796 (rank : 162) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRA_MOUSE
|
||||||
| NC score | 0.002516 (rank : 163) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18293 | Gene names | Npr1, Npra | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
ANPRA_HUMAN
|
||||||
| NC score | 0.002513 (rank : 164) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16066, Q6P4Q3 | Gene names | NPR1, ANPRA | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor A precursor (ANP-A) (ANPRA) (GC-A) (Guanylate cyclase) (EC 4.6.1.2) (NPR-A) (Atrial natriuretic peptide A-type receptor). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.002366 (rank : 165) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
MPP4_HUMAN
|
||||||
| NC score | 0.000872 (rank : 166) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
KIF1B_MOUSE
|
||||||
| NC score | 0.000653 (rank : 167) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
AT1A2_MOUSE
|
||||||
| NC score | 0.000521 (rank : 168) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PIE5 | Gene names | Atp1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium/potassium-transporting ATPase alpha-2 chain precursor (EC 3.6.3.9) (Sodium pump 2) (Na+/K+ ATPase 2) (Alpha(+)). | |||||
|
HRH1_HUMAN
|
||||||
| NC score | -0.001023 (rank : 169) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35367 | Gene names | HRH1 | |||
|
Domain Architecture |

|
|||||
| Description | Histamine H1 receptor. | |||||
|
LGR4_HUMAN
|
||||||
| NC score | -0.001168 (rank : 170) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXB1, Q9NYD1 | Gene names | LGR4, GPR48 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing G-protein coupled receptor 4 precursor (G-protein coupled receptor 48). | |||||
|
OLF9_MOUSE
|
||||||
| NC score | -0.001771 (rank : 171) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60885, Q8VGC0 | Gene names | Olfr9, Mor269-3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 9 (Olfactory receptor 269-3) (Odorant receptor M25). | |||||
|
OX2R_HUMAN
|
||||||
| NC score | -0.002590 (rank : 172) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43614 | Gene names | HCRTR2 | |||
|
Domain Architecture |

|
|||||
| Description | Orexin receptor type 2 (Ox2r) (Hypocretin receptor type 2). | |||||
|
OX2R_MOUSE
|
||||||
| NC score | -0.002616 (rank : 173) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P58308, Q5SGC8, Q6VLX3, Q8BG12 | Gene names | Hcrtr2, Mox2r | |||
|
Domain Architecture |

|
|||||
| Description | Orexin receptor type 2 (Ox2r) (Hypocretin receptor type 2). | |||||
|
KLF5_HUMAN
|
||||||
| NC score | -0.005019 (rank : 174) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13887, Q9UHP8 | Gene names | KLF5, BTEB2, CKLF, IKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Colon krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2) (GC box-binding protein 2). | |||||
|
ZN184_HUMAN
|
||||||
| NC score | -0.005540 (rank : 175) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 157 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99676, O60792, Q8TBA9 | Gene names | ZNF184 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 184. | |||||