Please be patient as the page loads
|
CASC5_MOUSE
|
||||||
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CASC5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.808324 (rank : 2) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
CASC5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
AF9_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 3) | NC score | 0.099203 (rank : 4) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
TENN_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.029726 (rank : 32) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Z71 | Gene names | Tnn | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
VEZA_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.100659 (rank : 3) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.039442 (rank : 18) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.056773 (rank : 6) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
NEIL3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.059019 (rank : 5) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.029228 (rank : 37) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.048384 (rank : 10) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.042470 (rank : 15) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.051996 (rank : 8) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.047820 (rank : 11) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.050027 (rank : 9) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
ACOT1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.024861 (rank : 43) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55137, Q549A9 | Gene names | Acot1, Cte1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 1 (EC 3.1.2.2) (Acyl-CoA thioesterase 1) (Inducible cytosolic acyl-coenzyme A thioester hydrolase) (Long chain acyl-CoA thioester hydrolase) (Long chain acyl-CoA hydrolase) (CTE-I). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.028070 (rank : 39) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CR034_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.054139 (rank : 7) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.041877 (rank : 16) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.032610 (rank : 26) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
CPIN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.047022 (rank : 12) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WTY4, Q3UJW5, Q8VC24, Q91W83 | Gene names | Ciapin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anamorsin (Cytokine-induced apoptosis inhibitor 1). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.020604 (rank : 49) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.045317 (rank : 13) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.037667 (rank : 21) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.021983 (rank : 47) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
EVC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.038656 (rank : 20) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57680 | Gene names | Evc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein homolog. | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.035670 (rank : 23) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
LHR2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.034526 (rank : 24) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.012644 (rank : 63) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
RP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.026707 (rank : 41) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |
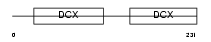
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.030062 (rank : 30) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
AHI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.014832 (rank : 61) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
AKA10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.036443 (rank : 22) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
CI084_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.038796 (rank : 19) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VXU9, Q96M73 | Gene names | C9orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf84. | |||||
|
KINH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.022891 (rank : 44) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.044558 (rank : 14) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.031544 (rank : 29) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.004905 (rank : 69) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
TLR2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.012010 (rank : 64) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60603, O15454, Q8NI00 | Gene names | TLR2, TIL4 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 2 precursor (Toll/interleukin 1 receptor-like protein 4) (CD282 antigen). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.033882 (rank : 25) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
ANGP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.022685 (rank : 45) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
APBA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.017456 (rank : 52) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |
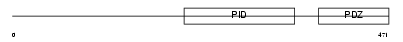
|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.021691 (rank : 48) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
NUF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.032419 (rank : 28) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.007084 (rank : 66) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.029969 (rank : 31) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
NEUT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.032597 (rank : 27) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D3P9 | Gene names | Nts | |||
|
Domain Architecture |
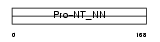
|
|||||
| Description | Neurotensin/neuromedin N precursor [Contains: Large neuromedin N (NmN- 125); Neuromedin N (NmN) (NN); Neurotensin (NT); Tail peptide]. | |||||
|
PININ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.040867 (rank : 17) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.028041 (rank : 40) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
COE3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.016912 (rank : 56) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
COE3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.016928 (rank : 55) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.022072 (rank : 46) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.008042 (rank : 65) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
NRIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.019775 (rank : 50) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
AKA10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.029259 (rank : 35) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
CD5R1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.016678 (rank : 57) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15078 | Gene names | CDK5R1, CDK5R, NCK5A | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
DPOLN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.017241 (rank : 54) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z5Q5, Q4TTW4, Q6ZNF4 | Gene names | POLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase nu (EC 2.7.7.7). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.029252 (rank : 36) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.015609 (rank : 59) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
LYPL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.018243 (rank : 51) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VWZ2, Q5VWZ3, Q7Z4A3, Q96AV0 | Gene names | LYPLAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysophospholipase-like protein 1 (EC 3.1.2.-). | |||||
|
MAT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.029214 (rank : 38) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51948, Q15817 | Gene names | MNAT1, CAP35, MAT1, RNF66 | |||
|
Domain Architecture |
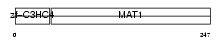
|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35) (Cyclin G1-interacting protein) (RING finger protein 66). | |||||
|
MAT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.029393 (rank : 34) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51949, Q3TZP0, Q9D8A0, Q9D8D2 | Gene names | Mnat1, Mat1 | |||
|
Domain Architecture |
|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35). | |||||
|
MPH6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.025152 (rank : 42) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99547 | Gene names | MPHOSPH6, MPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 6. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.015765 (rank : 58) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.029421 (rank : 33) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
ABCAA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.006536 (rank : 67) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWZ4, Q6PIQ6, Q7Z2I9, Q7Z7P7, Q86TD2 | Gene names | ABCA10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 10. | |||||
|
CDC5L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.017334 (rank : 53) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6A068, Q8K1J9 | Gene names | Cdc5l, Kiaa0432 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-related protein (Cdc5-like protein). | |||||
|
EMR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.004370 (rank : 70) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY15 | Gene names | EMR3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 3 precursor (EGF-like module-containing mucin-like receptor EMR3). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.014867 (rank : 60) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
SYTL5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.006100 (rank : 68) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
ZC3H5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.014124 (rank : 62) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
CASC5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
CASC5_HUMAN
|
||||||
| NC score | 0.808324 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
VEZA_HUMAN
|
||||||
| NC score | 0.100659 (rank : 3) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.099203 (rank : 4) | θ value | 0.00509761 (rank : 3) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
NEIL3_HUMAN
|
||||||
| NC score | 0.059019 (rank : 5) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TAT5, Q2PPJ3, Q8NG51, Q9NV95 | Gene names | NEIL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease VIII-like 3 (Nei-like 3) (DNA glycosylase FPG2). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.056773 (rank : 6) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CR034_HUMAN
|
||||||
| NC score | 0.054139 (rank : 7) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.051996 (rank : 8) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.050027 (rank : 9) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.048384 (rank : 10) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.047820 (rank : 11) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CPIN1_MOUSE
|
||||||
| NC score | 0.047022 (rank : 12) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WTY4, Q3UJW5, Q8VC24, Q91W83 | Gene names | Ciapin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anamorsin (Cytokine-induced apoptosis inhibitor 1). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.045317 (rank : 13) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.044558 (rank : 14) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.042470 (rank : 15) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.041877 (rank : 16) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.040867 (rank : 17) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.039442 (rank : 18) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CI084_HUMAN
|
||||||
| NC score | 0.038796 (rank : 19) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VXU9, Q96M73 | Gene names | C9orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf84. | |||||
|
EVC_MOUSE
|
||||||
| NC score | 0.038656 (rank : 20) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57680 | Gene names | Evc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ellis-van Creveld syndrome protein homolog. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.037667 (rank : 21) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
AKA10_MOUSE
|
||||||
| NC score | 0.036443 (rank : 22) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.035670 (rank : 23) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
LHR2A_HUMAN
|
||||||
| NC score | 0.034526 (rank : 24) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00534, Q6UN19, Q6UN20, Q9BVF8 | Gene names | LOH11CR2A, BCSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Loss of heterozygosity 11 chromosomal region 2 gene A protein (Breast cancer suppressor candidate 1) (BCSC-1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.033882 (rank : 25) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.032610 (rank : 26) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
NEUT_MOUSE
|
||||||
| NC score | 0.032597 (rank : 27) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D3P9 | Gene names | Nts | |||
|
Domain Architecture |
|
|||||
| Description | Neurotensin/neuromedin N precursor [Contains: Large neuromedin N (NmN- 125); Neuromedin N (NmN) (NN); Neurotensin (NT); Tail peptide]. | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.032419 (rank : 28) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.031544 (rank : 29) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.030062 (rank : 30) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.029969 (rank : 31) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
TENN_MOUSE
|
||||||
| NC score | 0.029726 (rank : 32) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Z71 | Gene names | Tnn | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.029421 (rank : 33) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
MAT1_MOUSE
|
||||||
| NC score | 0.029393 (rank : 34) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51949, Q3TZP0, Q9D8A0, Q9D8D2 | Gene names | Mnat1, Mat1 | |||
|
Domain Architecture |

|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35). | |||||
|
AKA10_HUMAN
|
||||||
| NC score | 0.029259 (rank : 35) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.029252 (rank : 36) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.029228 (rank : 37) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
MAT1_HUMAN
|
||||||
| NC score | 0.029214 (rank : 38) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P51948, Q15817 | Gene names | MNAT1, CAP35, MAT1, RNF66 | |||
|
Domain Architecture |

|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35) (Cyclin G1-interacting protein) (RING finger protein 66). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.028070 (rank : 39) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.028041 (rank : 40) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
RP1_MOUSE
|
||||||
| NC score | 0.026707 (rank : 41) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
MPH6_HUMAN
|
||||||
| NC score | 0.025152 (rank : 42) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99547 | Gene names | MPHOSPH6, MPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 6. | |||||
|
ACOT1_MOUSE
|
||||||
| NC score | 0.024861 (rank : 43) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O55137, Q549A9 | Gene names | Acot1, Cte1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A thioesterase 1 (EC 3.1.2.2) (Acyl-CoA thioesterase 1) (Inducible cytosolic acyl-coenzyme A thioester hydrolase) (Long chain acyl-CoA thioester hydrolase) (Long chain acyl-CoA hydrolase) (CTE-I). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.022891 (rank : 44) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
ANGP2_MOUSE
|
||||||
| NC score | 0.022685 (rank : 45) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35608, Q3U1A1, Q9D2D2 | Gene names | Angpt2, Agpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.022072 (rank : 46) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.021983 (rank : 47) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.021691 (rank : 48) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.020604 (rank : 49) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NRIP1_HUMAN
|
||||||
| NC score | 0.019775 (rank : 50) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48552, Q8IWE8 | Gene names | NRIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
LYPL1_HUMAN
|
||||||
| NC score | 0.018243 (rank : 51) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VWZ2, Q5VWZ3, Q7Z4A3, Q96AV0 | Gene names | LYPLAL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysophospholipase-like protein 1 (EC 3.1.2.-). | |||||
|
APBA3_MOUSE
|
||||||
| NC score | 0.017456 (rank : 52) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
CDC5L_MOUSE
|
||||||
| NC score | 0.017334 (rank : 53) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6A068, Q8K1J9 | Gene names | Cdc5l, Kiaa0432 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-related protein (Cdc5-like protein). | |||||
|
DPOLN_HUMAN
|
||||||
| NC score | 0.017241 (rank : 54) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z5Q5, Q4TTW4, Q6ZNF4 | Gene names | POLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase nu (EC 2.7.7.7). | |||||
|
COE3_MOUSE
|
||||||
| NC score | 0.016928 (rank : 55) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
COE3_HUMAN
|
||||||
| NC score | 0.016912 (rank : 56) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
CD5R1_HUMAN
|
||||||
| NC score | 0.016678 (rank : 57) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15078 | Gene names | CDK5R1, CDK5R, NCK5A | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.015765 (rank : 58) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.015609 (rank : 59) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.014867 (rank : 60) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
AHI1_HUMAN
|
||||||
| NC score | 0.014832 (rank : 61) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 537 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N157, Q504T3, Q5TCP9, Q6P098, Q6PIT6, Q8NDX0, Q9H0H2 | Gene names | AHI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Jouberin (Abelson helper integration site 1 protein homolog) (AHI-1). | |||||
|
ZC3H5_MOUSE
|
||||||
| NC score | 0.014124 (rank : 62) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.012644 (rank : 63) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
TLR2_HUMAN
|
||||||
| NC score | 0.012010 (rank : 64) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60603, O15454, Q8NI00 | Gene names | TLR2, TIL4 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 2 precursor (Toll/interleukin 1 receptor-like protein 4) (CD282 antigen). | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.008042 (rank : 65) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.007084 (rank : 66) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
ABCAA_HUMAN
|
||||||
| NC score | 0.006536 (rank : 67) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWZ4, Q6PIQ6, Q7Z2I9, Q7Z7P7, Q86TD2 | Gene names | ABCA10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 10. | |||||
|
SYTL5_MOUSE
|
||||||
| NC score | 0.006100 (rank : 68) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.004905 (rank : 69) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
EMR3_HUMAN
|
||||||
| NC score | 0.004370 (rank : 70) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY15 | Gene names | EMR3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 3 precursor (EGF-like module-containing mucin-like receptor EMR3). | |||||