Please be patient as the page loads
|
AKA10_MOUSE
|
||||||
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AKA10_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989098 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
SNX13_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 3) | NC score | 0.291625 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
RGS14_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 4) | NC score | 0.340628 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 5) | NC score | 0.352436 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 6) | NC score | 0.348571 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
SNX13_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 7) | NC score | 0.278060 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
RGS14_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 8) | NC score | 0.332487 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 9) | NC score | 0.245016 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS17_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 10) | NC score | 0.338925 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 11) | NC score | 0.218785 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS4_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.331826 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS17_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.335102 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |
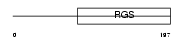
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 14) | NC score | 0.323382 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |
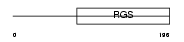
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 15) | NC score | 0.321571 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |
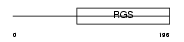
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 16) | NC score | 0.286869 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS13_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 17) | NC score | 0.332229 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |
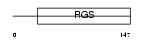
|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS13_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 18) | NC score | 0.328211 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS20_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 19) | NC score | 0.326504 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS20_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.320387 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
SNX14_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.163551 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
SNX14_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 22) | NC score | 0.167921 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
RGS16_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.306382 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS8_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.310273 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.025355 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
RGS5_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.308769 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS19_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.310314 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS8_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.307549 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS10_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.308143 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.056029 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.088277 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
EP15_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.042990 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
RGS19_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.300784 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS10_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.301707 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
4ET_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.067688 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
4ET_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.067396 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.041898 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
AXN1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.227169 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
PRD16_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.004563 (rank : 84) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.033837 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.037446 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CA082_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.042989 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IXW5, Q49AS7, Q9H8Y2 | Gene names | C1orf82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0385 protein C1orf82. | |||||
|
A1AT5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.018960 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00898 | Gene names | Serpina1e, Dom5, Spi1-5 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-antitrypsin 1-5 precursor (Serine protease inhibitor 1-5) (Alpha-1 protease inhibitor 5). | |||||
|
RGS16_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.285583 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.049144 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.012133 (rank : 81) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.026384 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.192497 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
RGS5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.287807 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
CASC5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.036443 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
CTNA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.021238 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26231 | Gene names | Ctnna1, Catna1 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-1 (102 kDa cadherin-associated protein) (CAP102) (Alpha E-catenin). | |||||
|
EPC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.028167 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.039702 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.025570 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
AXN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.195307 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.025119 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
ADAM8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.005421 (rank : 82) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05910 | Gene names | Adam8, Ms2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (Macrophage cysteine-rich glycoprotein) (CD156 antigen). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.023352 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
A1AT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.016351 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07758, Q80YB8, Q8JZV6, Q91XB8 | Gene names | Serpina1a, Dom1, Spi1-1 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-antitrypsin 1-1 precursor (Serine protease inhibitor 1-1) (Alpha-1 protease inhibitor 1) (Alpha-1-antiproteinase) (AAT). | |||||
|
A1AT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.016636 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22599, Q61283, Q80ZH5, Q8VC20 | Gene names | Serpina1b, Aat2, Dom2, Spi1-2 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-antitrypsin 1-2 precursor (Serine protease inhibitor 1-2) (Alpha-1 protease inhibitor 2) (Alpha-1-antiproteinase) (AAT). | |||||
|
A1AT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.016654 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00896 | Gene names | Serpina1c, Dom3, Spi1-3 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1-antitrypsin 1-3 precursor (Serine protease inhibitor 1-3) (Alpha-1 protease inhibitor 3). | |||||
|
A1AT6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.016676 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P81105, Q91V74, Q91WH5, Q91XC1 | Gene names | Spi1-6, Dom6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1-antitrypsin 1-6 precursor (Serine protease inhibitor 1-6) (Alpha-1 protease inhibitor 6). | |||||
|
CTNA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.018787 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35221, Q12795 | Gene names | CTNNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-1 (Cadherin-associated protein) (Alpha E-catenin) (NY- REN-13 antigen). | |||||
|
EST1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.026345 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.019310 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
RGS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.285407 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.016988 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.023054 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.017055 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
EPC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.023477 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0I4, Q3URX9, Q80Y70, Q8BXC5 | Gene names | Epc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.002093 (rank : 85) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.004891 (rank : 83) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.023886 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
K0226_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.013288 (rank : 80) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.001842 (rank : 86) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
AXN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.196854 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
RGS11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.206377 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |
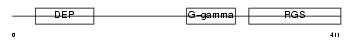
|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.259109 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |
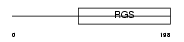
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS18_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.259911 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |
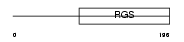
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.181050 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.191947 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.197597 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |
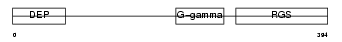
|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.197265 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.191131 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.190855 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
SNX25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.092402 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
AKA10_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_HUMAN
|
||||||
| NC score | 0.989098 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
RGS1_HUMAN
|
||||||
| NC score | 0.352436 (rank : 3) | θ value | 7.1131e-05 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS1_MOUSE
|
||||||
| NC score | 0.348571 (rank : 4) | θ value | 0.00020696 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS14_HUMAN
|
||||||
| NC score | 0.340628 (rank : 5) | θ value | 7.1131e-05 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS17_HUMAN
|
||||||
| NC score | 0.338925 (rank : 6) | θ value | 0.000461057 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS17_MOUSE
|
||||||
| NC score | 0.335102 (rank : 7) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS14_MOUSE
|
||||||
| NC score | 0.332487 (rank : 8) | θ value | 0.000270298 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS13_HUMAN
|
||||||
| NC score | 0.332229 (rank : 9) | θ value | 0.00390308 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS4_HUMAN
|
||||||
| NC score | 0.331826 (rank : 10) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS13_MOUSE
|
||||||
| NC score | 0.328211 (rank : 11) | θ value | 0.00390308 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS20_MOUSE
|
||||||
| NC score | 0.326504 (rank : 12) | θ value | 0.00665767 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS2_HUMAN
|
||||||
| NC score | 0.323382 (rank : 13) | θ value | 0.00228821 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS2_MOUSE
|
||||||
| NC score | 0.321571 (rank : 14) | θ value | 0.00298849 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS20_HUMAN
|
||||||
| NC score | 0.320387 (rank : 15) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS19_MOUSE
|
||||||
| NC score | 0.310314 (rank : 16) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS8_HUMAN
|
||||||
| NC score | 0.310273 (rank : 17) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS5_MOUSE
|
||||||
| NC score | 0.308769 (rank : 18) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS10_MOUSE
|
||||||
| NC score | 0.308143 (rank : 19) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS8_MOUSE
|
||||||
| NC score | 0.307549 (rank : 20) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS16_MOUSE
|
||||||
| NC score | 0.306382 (rank : 21) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS10_HUMAN
|
||||||
| NC score | 0.301707 (rank : 22) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS19_HUMAN
|
||||||
| NC score | 0.300784 (rank : 23) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
SNX13_MOUSE
|
||||||
| NC score | 0.291625 (rank : 24) | θ value | 1.87187e-05 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
RGS5_HUMAN
|
||||||
| NC score | 0.287807 (rank : 25) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.286869 (rank : 26) | θ value | 0.00390308 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS16_HUMAN
|
||||||
| NC score | 0.285583 (rank : 27) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS4_MOUSE
|
||||||
| NC score | 0.285407 (rank : 28) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
SNX13_HUMAN
|
||||||
| NC score | 0.278060 (rank : 29) | θ value | 0.00020696 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
RGS18_MOUSE
|
||||||
| NC score | 0.259911 (rank : 30) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS18_HUMAN
|
||||||
| NC score | 0.259109 (rank : 31) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.245016 (rank : 32) | θ value | 0.00035302 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
AXN1_MOUSE
|
||||||
| NC score | 0.227169 (rank : 33) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.218785 (rank : 34) | θ value | 0.000602161 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS11_HUMAN
|
||||||
| NC score | 0.206377 (rank : 35) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS7_HUMAN
|
||||||
| NC score | 0.197597 (rank : 36) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| NC score | 0.197265 (rank : 37) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
AXN1_HUMAN
|
||||||
| NC score | 0.196854 (rank : 38) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.195307 (rank : 39) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.192497 (rank : 40) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
RGS6_MOUSE
|
||||||
| NC score | 0.191947 (rank : 41) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS9_HUMAN
|
||||||
| NC score | 0.191131 (rank : 42) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| NC score | 0.190855 (rank : 43) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS6_HUMAN
|
||||||
| NC score | 0.181050 (rank : 44) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
SNX14_MOUSE
|
||||||
| NC score | 0.167921 (rank : 45) | θ value | 0.0252991 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
SNX14_HUMAN
|
||||||
| NC score | 0.163551 (rank : 46) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
SNX25_HUMAN
|
||||||
| NC score | 0.092402 (rank : 47) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.088277 (rank : 48) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
4ET_HUMAN
|
||||||
| NC score | 0.067688 (rank : 49) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
4ET_MOUSE
|
||||||
| NC score | 0.067396 (rank : 50) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.056029 (rank : 51) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.049144 (rank : 52) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.042990 (rank : 53) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
CA082_HUMAN
|
||||||
| NC score | 0.042989 (rank : 54) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IXW5, Q49AS7, Q9H8Y2 | Gene names | C1orf82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0385 protein C1orf82. | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.041898 (rank : 55) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.039702 (rank : 56) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.037446 (rank : 57) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CASC5_MOUSE
|
||||||
| NC score | 0.036443 (rank : 58) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.033837 (rank : 59) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
EPC2_HUMAN
|
||||||
| NC score | 0.028167 (rank : 60) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.026384 (rank : 61) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
EST1A_HUMAN
|
||||||
| NC score | 0.026345 (rank : 62) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.025570 (rank : 63) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.025355 (rank : 64) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.025119 (rank : 65) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.023886 (rank : 66) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
EPC2_MOUSE
|
||||||
| NC score | 0.023477 (rank : 67) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0I4, Q3URX9, Q80Y70, Q8BXC5 | Gene names | Epc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.023352 (rank : 68) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.023054 (rank : 69) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
CTNA1_MOUSE
|
||||||
| NC score | 0.021238 (rank : 70) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26231 | Gene names | Ctnna1, Catna1 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-1 (102 kDa cadherin-associated protein) (CAP102) (Alpha E-catenin). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.019310 (rank : 71) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
A1AT5_MOUSE
|
||||||
| NC score | 0.018960 (rank : 72) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00898 | Gene names | Serpina1e, Dom5, Spi1-5 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-5 precursor (Serine protease inhibitor 1-5) (Alpha-1 protease inhibitor 5). | |||||
|
CTNA1_HUMAN
|
||||||
| NC score | 0.018787 (rank : 73) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35221, Q12795 | Gene names | CTNNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin alpha-1 (Cadherin-associated protein) (Alpha E-catenin) (NY- REN-13 antigen). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.017055 (rank : 74) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.016988 (rank : 75) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
A1AT6_MOUSE
|
||||||
| NC score | 0.016676 (rank : 76) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P81105, Q91V74, Q91WH5, Q91XC1 | Gene names | Spi1-6, Dom6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1-antitrypsin 1-6 precursor (Serine protease inhibitor 1-6) (Alpha-1 protease inhibitor 6). | |||||
|
A1AT3_MOUSE
|
||||||
| NC score | 0.016654 (rank : 77) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00896 | Gene names | Serpina1c, Dom3, Spi1-3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-3 precursor (Serine protease inhibitor 1-3) (Alpha-1 protease inhibitor 3). | |||||
|
A1AT2_MOUSE
|
||||||
| NC score | 0.016636 (rank : 78) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22599, Q61283, Q80ZH5, Q8VC20 | Gene names | Serpina1b, Aat2, Dom2, Spi1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-2 precursor (Serine protease inhibitor 1-2) (Alpha-1 protease inhibitor 2) (Alpha-1-antiproteinase) (AAT). | |||||
|
A1AT1_MOUSE
|
||||||
| NC score | 0.016351 (rank : 79) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07758, Q80YB8, Q8JZV6, Q91XB8 | Gene names | Serpina1a, Dom1, Spi1-1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-antitrypsin 1-1 precursor (Serine protease inhibitor 1-1) (Alpha-1 protease inhibitor 1) (Alpha-1-antiproteinase) (AAT). | |||||
|
K0226_HUMAN
|
||||||
| NC score | 0.013288 (rank : 80) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92622 | Gene names | KIAA0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.012133 (rank : 81) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ADAM8_MOUSE
|
||||||
| NC score | 0.005421 (rank : 82) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05910 | Gene names | Adam8, Ms2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (Macrophage cysteine-rich glycoprotein) (CD156 antigen). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.004891 (rank : 83) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
PRD16_HUMAN
|
||||||
| NC score | 0.004563 (rank : 84) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HAZ2, Q8WYJ9, Q9C0I8 | Gene names | PRDM16, KIAA1675, MEL1, PFM13 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 16 (PR domain-containing protein 16) (Transcription factor MEL1). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.002093 (rank : 85) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.001842 (rank : 86) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||