Please be patient as the page loads
|
AXN2_HUMAN
|
||||||
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AXN2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989516 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN1_MOUSE
|
||||||
| θ value | 6.38546e-147 (rank : 3) | NC score | 0.954450 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN1_HUMAN
|
||||||
| θ value | 8.33966e-147 (rank : 4) | NC score | 0.953621 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
RGS18_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 5) | NC score | 0.687936 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS18_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 6) | NC score | 0.686424 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 7) | NC score | 0.491598 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 8) | NC score | 0.435760 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS10_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 9) | NC score | 0.691038 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS2_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 10) | NC score | 0.671314 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS1_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 11) | NC score | 0.667241 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |
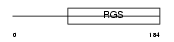
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS2_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 12) | NC score | 0.667182 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |
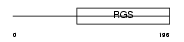
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS1_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 13) | NC score | 0.670805 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |
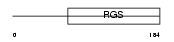
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS10_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 14) | NC score | 0.683127 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |
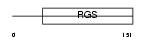
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS8_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 15) | NC score | 0.668310 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS8_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 16) | NC score | 0.667487 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS4_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 17) | NC score | 0.663090 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS5_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 18) | NC score | 0.669426 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS4_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 19) | NC score | 0.662454 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS14_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 20) | NC score | 0.618771 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS5_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 21) | NC score | 0.662722 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS13_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 22) | NC score | 0.655451 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS14_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 23) | NC score | 0.608491 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS16_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 24) | NC score | 0.651537 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS19_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 25) | NC score | 0.634855 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS16_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 26) | NC score | 0.654085 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS20_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 27) | NC score | 0.633363 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS13_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 28) | NC score | 0.656305 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS17_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 29) | NC score | 0.636013 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS19_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 30) | NC score | 0.632871 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS17_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 31) | NC score | 0.627288 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS20_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 32) | NC score | 0.620860 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS11_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 33) | NC score | 0.557248 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 34) | NC score | 0.513123 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS9_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 35) | NC score | 0.536527 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 36) | NC score | 0.537105 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |
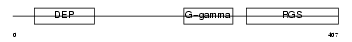
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 37) | NC score | 0.238175 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 38) | NC score | 0.237756 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 39) | NC score | 0.245908 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
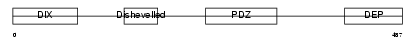
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 40) | NC score | 0.246209 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL1L_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 41) | NC score | 0.220924 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |
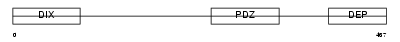
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
DVL1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 42) | NC score | 0.218633 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |
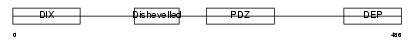
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
DVL1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 43) | NC score | 0.218090 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
RGS6_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 44) | NC score | 0.491172 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS7_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 45) | NC score | 0.504767 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |
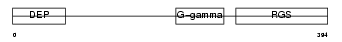
|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 46) | NC score | 0.497885 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
LMBTL_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 47) | NC score | 0.038704 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
RGS6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 48) | NC score | 0.462805 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 49) | NC score | 0.029760 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.048295 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
AAKG2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.025753 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WG5, Q6V7V5 | Gene names | Prkag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.048311 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.040401 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.014572 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TEAD2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.028122 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15562, Q96IG3 | Gene names | TEAD2, TEF4 | |||
|
Domain Architecture |
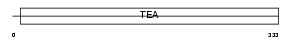
|
|||||
| Description | Transcriptional enhancer factor TEF-4 (TEA domain family member 2) (TEAD-2). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.039045 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.024797 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.001738 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.030960 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
KIF1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.010556 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12756 | Gene names | KIF1A, ATSV | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
KIF1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.012004 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33173, Q61770 | Gene names | Kif1a, Atsv, Kif1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
PMS2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.020073 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PARD3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.038212 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
TEAD2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.022375 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48301, Q62297 | Gene names | Tead2, Etdf, Etf, Tef4 | |||
|
Domain Architecture |
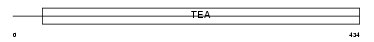
|
|||||
| Description | Transcriptional enhancer factor TEF-4 (TEA domain family member 2) (TEAD-2) (Embryonic TEA domain-containing factor) (ETF) (ETEF-1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.014419 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PUNC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.006431 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IVU1, O95215 | Gene names | PUNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
SEN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.021801 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P7W5, Q3UYG6 | Gene names | Tsen2, Sen2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen2 (EC 3.1.27.9) (tRNA-intron endonuclease Sen2). | |||||
|
AKA10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.195746 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.195307 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.007557 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CV106_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.036429 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P0N0, Q86V14, Q96PY4, Q9NUR5, Q9Y4X9 | Gene names | C14orf106, KIAA1903 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf106 (P243). | |||||
|
MYPC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.008654 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14324 | Gene names | MYBPC2, MYBPCF | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
SCG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.029678 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
SNX14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.089893 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
SNX14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.088460 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
5NT1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.017998 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P26, Q8N9W3, Q8NA26, Q96DU5, Q96KE6, Q96M25, Q96SA3 | Gene names | NT5C1B, AIRP | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.012606 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
CT112_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.026017 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MY1, Q5JYB7, Q6P0Y4, Q9BR34, Q9NQF6 | Gene names | C20orf112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf112. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.018262 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GASP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.009947 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GCC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.005165 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
GCC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.006195 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4H2, Q8CCR3 | Gene names | Gcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.008213 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ITPR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.008970 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
MYPC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.007879 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5XKE0, Q8C109, Q8K2V0 | Gene names | Mybpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.005212 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
ULK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | -0.000502 (rank : 105) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||
|
ACF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.004923 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.016900 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
ICOSL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.009664 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHJ8 | Gene names | Icoslg, B7h2, B7rp1, Icosl | |||
|
Domain Architecture |
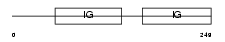
|
|||||
| Description | ICOS ligand precursor (B7 homolog 2) (B7-H2) (B7-like protein Gl50) (B7-related protein 1) (B7RP-1) (LICOS) (CD275 antigen). | |||||
|
KBTB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.001942 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHI4, Q8BNJ8 | Gene names | Kbtbd3, Bklhd3 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 3 (BTB and kelch domain-containing protein 3). | |||||
|
PAX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.005405 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09084, Q9R2B1 | Gene names | Pax1, Pax-1 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-1. | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.006129 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.009043 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
AN13D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.005425 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZTN6, Q6ZVD0, Q86SU1 | Gene names | ANKRD13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13D. | |||||
|
CNTP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.002315 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
GPC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.005056 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKV1 | Gene names | Gpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-2 precursor. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.012801 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.006146 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.016902 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.014825 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
WDR4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.007443 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EP82 | Gene names | Wdr4 | |||
|
Domain Architecture |
|
|||||
| Description | WD repeat protein 4. | |||||
|
SNX13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.165306 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SNX13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.167430 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
SNX25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.075652 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.989516 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN1_MOUSE
|
||||||
| NC score | 0.954450 (rank : 3) | θ value | 6.38546e-147 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN1_HUMAN
|
||||||
| NC score | 0.953621 (rank : 4) | θ value | 8.33966e-147 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
RGS10_HUMAN
|
||||||
| NC score | 0.691038 (rank : 5) | θ value | 1.02475e-11 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS18_HUMAN
|
||||||
| NC score | 0.687936 (rank : 6) | θ value | 1.52774e-15 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS18_MOUSE
|
||||||
| NC score | 0.686424 (rank : 7) | θ value | 1.68911e-14 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS10_MOUSE
|
||||||
| NC score | 0.683127 (rank : 8) | θ value | 6.64225e-11 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS2_HUMAN
|
||||||
| NC score | 0.671314 (rank : 9) | θ value | 1.02475e-11 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS1_HUMAN
|
||||||
| NC score | 0.670805 (rank : 10) | θ value | 5.08577e-11 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS5_HUMAN
|
||||||
| NC score | 0.669426 (rank : 11) | θ value | 7.34386e-10 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS8_MOUSE
|
||||||
| NC score | 0.668310 (rank : 12) | θ value | 6.64225e-11 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS8_HUMAN
|
||||||
| NC score | 0.667487 (rank : 13) | θ value | 8.67504e-11 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS1_MOUSE
|
||||||
| NC score | 0.667241 (rank : 14) | θ value | 3.89403e-11 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS2_MOUSE
|
||||||
| NC score | 0.667182 (rank : 15) | θ value | 3.89403e-11 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS4_HUMAN
|
||||||
| NC score | 0.663090 (rank : 16) | θ value | 2.52405e-10 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS5_MOUSE
|
||||||
| NC score | 0.662722 (rank : 17) | θ value | 4.76016e-09 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS4_MOUSE
|
||||||
| NC score | 0.662454 (rank : 18) | θ value | 1.25267e-09 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS13_MOUSE
|
||||||
| NC score | 0.656305 (rank : 19) | θ value | 8.97725e-08 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS13_HUMAN
|
||||||
| NC score | 0.655451 (rank : 20) | θ value | 8.11959e-09 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS16_MOUSE
|
||||||
| NC score | 0.654085 (rank : 21) | θ value | 3.08544e-08 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS16_HUMAN
|
||||||
| NC score | 0.651537 (rank : 22) | θ value | 1.38499e-08 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS17_MOUSE
|
||||||
| NC score | 0.636013 (rank : 23) | θ value | 1.17247e-07 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS19_MOUSE
|
||||||
| NC score | 0.634855 (rank : 24) | θ value | 1.38499e-08 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS20_MOUSE
|
||||||
| NC score | 0.633363 (rank : 25) | θ value | 3.08544e-08 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS19_HUMAN
|
||||||
| NC score | 0.632871 (rank : 26) | θ value | 1.17247e-07 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS17_HUMAN
|
||||||
| NC score | 0.627288 (rank : 27) | θ value | 1.99992e-07 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS20_HUMAN
|
||||||
| NC score | 0.620860 (rank : 28) | θ value | 9.92553e-07 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS14_MOUSE
|
||||||
| NC score | 0.618771 (rank : 29) | θ value | 2.79066e-09 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS14_HUMAN
|
||||||
| NC score | 0.608491 (rank : 30) | θ value | 1.38499e-08 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS11_HUMAN
|
||||||
| NC score | 0.557248 (rank : 31) | θ value | 1.69304e-06 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS9_MOUSE
|
||||||
| NC score | 0.537105 (rank : 32) | θ value | 1.43324e-05 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_HUMAN
|
||||||
| NC score | 0.536527 (rank : 33) | θ value | 1.43324e-05 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.513123 (rank : 34) | θ value | 1.43324e-05 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS7_HUMAN
|
||||||
| NC score | 0.504767 (rank : 35) | θ value | 0.00134147 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| NC score | 0.497885 (rank : 36) | θ value | 0.00298849 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.491598 (rank : 37) | θ value | 2.43908e-13 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS6_MOUSE
|
||||||
| NC score | 0.491172 (rank : 38) | θ value | 0.00102713 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS6_HUMAN
|
||||||
| NC score | 0.462805 (rank : 39) | θ value | 0.0193708 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.435760 (rank : 40) | θ value | 6.00763e-12 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.246209 (rank : 41) | θ value | 1.87187e-05 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.245908 (rank : 42) | θ value | 1.87187e-05 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.238175 (rank : 43) | θ value | 1.87187e-05 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.237756 (rank : 44) | θ value | 1.87187e-05 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL1L_HUMAN
|
||||||
| NC score | 0.220924 (rank : 45) | θ value | 0.000602161 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
DVL1_MOUSE
|
||||||
| NC score | 0.218633 (rank : 46) | θ value | 0.000602161 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
DVL1_HUMAN
|
||||||
| NC score | 0.218090 (rank : 47) | θ value | 0.000786445 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
AKA10_HUMAN
|
||||||
| NC score | 0.195746 (rank : 48) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| NC score | 0.195307 (rank : 49) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
SNX13_MOUSE
|
||||||
| NC score | 0.167430 (rank : 50) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
SNX13_HUMAN
|
||||||
| NC score | 0.165306 (rank : 51) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SNX14_HUMAN
|
||||||
| NC score | 0.089893 (rank : 52) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
SNX14_MOUSE
|
||||||
| NC score | 0.088460 (rank : 53) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
SNX25_HUMAN
|
||||||
| NC score | 0.075652 (rank : 54) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.048311 (rank : 55) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.048295 (rank : 56) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.040401 (rank : 57) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.039045 (rank : 58) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
LMBTL_HUMAN
|
||||||
| NC score | 0.038704 (rank : 59) | θ value | 0.0148317 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
PARD3_MOUSE
|
||||||
| NC score | 0.038212 (rank : 60) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
CV106_HUMAN
|
||||||
| NC score | 0.036429 (rank : 61) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P0N0, Q86V14, Q96PY4, Q9NUR5, Q9Y4X9 | Gene names | C14orf106, KIAA1903 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf106 (P243). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.030960 (rank : 62) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.029760 (rank : 63) | θ value | 0.0563607 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SCG2_HUMAN
|
||||||
| NC score | 0.029678 (rank : 64) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
TEAD2_HUMAN
|
||||||
| NC score | 0.028122 (rank : 65) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15562, Q96IG3 | Gene names | TEAD2, TEF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional enhancer factor TEF-4 (TEA domain family member 2) (TEAD-2). | |||||
|
CT112_HUMAN
|
||||||
| NC score | 0.026017 (rank : 66) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MY1, Q5JYB7, Q6P0Y4, Q9BR34, Q9NQF6 | Gene names | C20orf112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf112. | |||||
|
AAKG2_MOUSE
|
||||||
| NC score | 0.025753 (rank : 67) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WG5, Q6V7V5 | Gene names | Prkag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.024797 (rank : 68) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
TEAD2_MOUSE
|
||||||
| NC score | 0.022375 (rank : 69) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48301, Q62297 | Gene names | Tead2, Etdf, Etf, Tef4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional enhancer factor TEF-4 (TEA domain family member 2) (TEAD-2) (Embryonic TEA domain-containing factor) (ETF) (ETEF-1). | |||||
|
SEN2_MOUSE
|
||||||
| NC score | 0.021801 (rank : 70) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P7W5, Q3UYG6 | Gene names | Tsen2, Sen2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen2 (EC 3.1.27.9) (tRNA-intron endonuclease Sen2). | |||||
|
PMS2_HUMAN
|
||||||
| NC score | 0.020073 (rank : 71) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.018262 (rank : 72) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
5NT1B_HUMAN
|
||||||
| NC score | 0.017998 (rank : 73) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P26, Q8N9W3, Q8NA26, Q96DU5, Q96KE6, Q96M25, Q96SA3 | Gene names | NT5C1B, AIRP | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.016902 (rank : 74) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.016900 (rank : 75) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.014825 (rank : 76) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.014572 (rank : 77) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.014419 (rank : 78) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.012801 (rank : 79) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.012606 (rank : 80) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
KIF1A_MOUSE
|
||||||
| NC score | 0.012004 (rank : 81) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33173, Q61770 | Gene names | Kif1a, Atsv, Kif1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
KIF1A_HUMAN
|
||||||
| NC score | 0.010556 (rank : 82) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12756 | Gene names | KIF1A, ATSV | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
GASP1_MOUSE
|
||||||
| NC score | 0.009947 (rank : 83) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
ICOSL_MOUSE
|
||||||
| NC score | 0.009664 (rank : 84) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHJ8 | Gene names | Icoslg, B7h2, B7rp1, Icosl | |||
|
Domain Architecture |

|
|||||
| Description | ICOS ligand precursor (B7 homolog 2) (B7-H2) (B7-like protein Gl50) (B7-related protein 1) (B7RP-1) (LICOS) (CD275 antigen). | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.009043 (rank : 85) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ITPR3_MOUSE
|
||||||
| NC score | 0.008970 (rank : 86) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70227, Q5DWM4, Q8CED5, Q91Z08 | Gene names | Itpr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 3 (Type 3 inositol 1,4,5- trisphosphate receptor) (Type 3 InsP3 receptor) (IP3 receptor isoform 3) (InsP3R3). | |||||
|
MYPC2_HUMAN
|
||||||
| NC score | 0.008654 (rank : 87) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14324 | Gene names | MYBPC2, MYBPCF | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.008213 (rank : 88) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MYPC2_MOUSE
|
||||||
| NC score | 0.007879 (rank : 89) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5XKE0, Q8C109, Q8K2V0 | Gene names | Mybpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-binding protein C, fast-type (Fast MyBP-C) (C-protein, skeletal muscle fast isoform). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.007557 (rank : 90) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
WDR4_MOUSE
|
||||||
| NC score | 0.007443 (rank : 91) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EP82 | Gene names | Wdr4 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 4. | |||||
|
PUNC_HUMAN
|
||||||
| NC score | 0.006431 (rank : 92) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IVU1, O95215 | Gene names | PUNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
GCC1_MOUSE
|
||||||
| NC score | 0.006195 (rank : 93) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4H2, Q8CCR3 | Gene names | Gcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.006146 (rank : 94) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.006129 (rank : 95) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
AN13D_HUMAN
|
||||||
| NC score | 0.005425 (rank : 96) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZTN6, Q6ZVD0, Q86SU1 | Gene names | ANKRD13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 13D. | |||||
|
PAX1_MOUSE
|
||||||
| NC score | 0.005405 (rank : 97) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09084, Q9R2B1 | Gene names | Pax1, Pax-1 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-1. | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.005212 (rank : 98) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
GCC1_HUMAN
|
||||||
| NC score | 0.005165 (rank : 99) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
GPC2_MOUSE
|
||||||
| NC score | 0.005056 (rank : 100) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKV1 | Gene names | Gpc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-2 precursor. | |||||
|
ACF_MOUSE
|
||||||
| NC score | 0.004923 (rank : 101) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5YD48, Q5FW55, Q5YD47 | Gene names | Acf, Asp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | APOBEC1 complementation factor (APOBEC1-stimulating protein). | |||||
|
CNTP2_MOUSE
|
||||||
| NC score | 0.002315 (rank : 102) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPW0, Q6P2K4, Q6ZQ31 | Gene names | Cntnap2, Kiaa0868 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-associated protein-like 2 precursor (Cell recognition molecule Caspr2). | |||||
|
KBTB3_MOUSE
|
||||||
| NC score | 0.001942 (rank : 103) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHI4, Q8BNJ8 | Gene names | Kbtbd3, Bklhd3 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 3 (BTB and kelch domain-containing protein 3). | |||||
|
ULK2_HUMAN
|
||||||
| NC score | 0.001738 (rank : 104) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
ULK2_MOUSE
|
||||||
| NC score | -0.000502 (rank : 105) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 943 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QY01, Q80TV7, Q9WTP4 | Gene names | Ulk2, Kiaa0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2) (Serine/threonine-protein kinase Unc51.2). | |||||