Please be patient as the page loads
|
RGS7_MOUSE
|
||||||
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RGS6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991056 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995354 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS7_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999572 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |
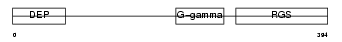
|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS9_HUMAN
|
||||||
| θ value | 8.77142e-88 (rank : 5) | NC score | 0.956928 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |
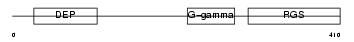
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| θ value | 5.6855e-87 (rank : 6) | NC score | 0.956417 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |
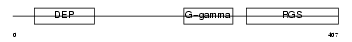
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS11_HUMAN
|
||||||
| θ value | 2.47191e-82 (rank : 7) | NC score | 0.963996 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |
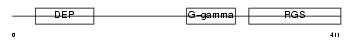
|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS20_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 8) | NC score | 0.856927 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS17_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 9) | NC score | 0.855553 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS5_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 10) | NC score | 0.854676 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS20_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 11) | NC score | 0.855878 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS19_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 12) | NC score | 0.854721 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |
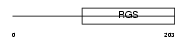
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS17_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 13) | NC score | 0.853260 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |
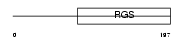
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS19_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 14) | NC score | 0.854066 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |
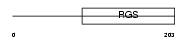
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS1_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 15) | NC score | 0.854554 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |
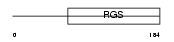
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS5_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 16) | NC score | 0.848032 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |
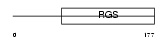
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS8_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 17) | NC score | 0.844342 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |
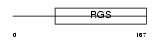
|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS8_MOUSE
|
||||||
| θ value | 2.43343e-21 (rank : 18) | NC score | 0.843314 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS18_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 19) | NC score | 0.847210 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |
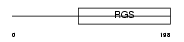
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS1_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 20) | NC score | 0.854807 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |
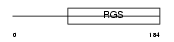
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 21) | NC score | 0.531297 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 22) | NC score | 0.595529 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS2_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 23) | NC score | 0.838651 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS2_MOUSE
|
||||||
| θ value | 1.2077e-20 (rank : 24) | NC score | 0.837463 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 25) | NC score | 0.717752 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS16_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 26) | NC score | 0.838064 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS4_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 27) | NC score | 0.837204 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS18_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 28) | NC score | 0.840938 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS4_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 29) | NC score | 0.839357 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS14_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 30) | NC score | 0.796584 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS13_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 31) | NC score | 0.840586 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS14_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 32) | NC score | 0.797139 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS10_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 33) | NC score | 0.850592 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS16_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 34) | NC score | 0.832421 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |
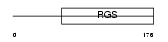
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS13_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 35) | NC score | 0.836049 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS10_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 36) | NC score | 0.849691 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |
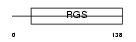
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
SNX25_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 37) | NC score | 0.243531 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 38) | NC score | 0.493758 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 39) | NC score | 0.509671 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 40) | NC score | 0.497885 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
SNX13_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 41) | NC score | 0.284449 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SNX13_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 42) | NC score | 0.285252 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 43) | NC score | 0.079306 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.074941 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
PREX1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 45) | NC score | 0.051004 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
AXN1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 46) | NC score | 0.491638 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
GBG7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.074863 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60262 | Gene names | GNG7, GNGT7 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-7 subunit precursor. | |||||
|
ABLM3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 48) | NC score | 0.014137 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94929, Q658S1, Q68CI5, Q9BV32 | Gene names | ABLIM3, KIAA0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.074244 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
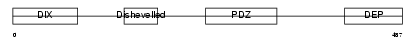
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.074307 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
GBG4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.086164 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50153 | Gene names | Gng4, Gngt4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-4 subunit precursor. | |||||
|
PLEK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.060454 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.077409 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
GBG12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.061023 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UBI6, Q9BRV5 | Gene names | GNG12 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-12 subunit precursor. | |||||
|
GBG2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.089599 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59768, Q6P9A9 | Gene names | GNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit precursor (G gamma-I). | |||||
|
GBG2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.089599 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63213, P16874, Q3TYE8, Q61013, Q9TS47 | Gene names | Gng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit precursor (G gamma-I). | |||||
|
GBG4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.081453 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50150 | Gene names | GNG4, GNGT4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-4 subunit precursor. | |||||
|
GBG7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.059142 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61016, Q8JZP6 | Gene names | Gng7, Gngt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-7 subunit precursor. | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.054152 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.053259 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.016425 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.016539 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
GBG12_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.059165 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DAS9, Q544P3 | Gene names | Gng12 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-12 subunit precursor. | |||||
|
GBG3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.070581 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63215, P29798, Q61014 | Gene names | GNG3, GNGT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-3 subunit precursor. | |||||
|
GBG3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.070581 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63216, P29798, Q61014 | Gene names | Gng3, Gngt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-3 subunit precursor. | |||||
|
GBG8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.079036 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UK08 | Gene names | GNG8, GNG9, GNGT9 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-8 subunit precursor (Gamma-9). | |||||
|
GBG8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.078994 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63078, P43426 | Gene names | Gng8, Gng9, Gngt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-8 subunit precursor (Gamma-9). | |||||
|
SYEP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.029874 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.002130 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.002079 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
RK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | -0.002173 (rank : 81) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15835 | Gene names | GRK1, RHOK | |||
|
Domain Architecture |

|
|||||
| Description | Rhodopsin kinase precursor (EC 2.7.11.14) (RK) (G protein-coupled receptor kinase 1). | |||||
|
SNX14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.053786 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
SNX14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.064207 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.013692 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
GAMT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.017274 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35969, Q3TZ58, Q3US90 | Gene names | Gamt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanidinoacetate N-methyltransferase (EC 2.1.1.2). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.000161 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.008035 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SMRD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.008508 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96GM5, Q92924, Q9Y635 | Gene names | SMARCD1, BAF60A | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A). | |||||
|
SMRD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.008508 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61466, P70384, Q8R0I7 | Gene names | Smarcd1, Baf60a, D15Kz1 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A) (D15KZ1 protein). | |||||
|
AKA10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.197709 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.197265 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
RGS7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_HUMAN
|
||||||
| NC score | 0.999572 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS6_MOUSE
|
||||||
| NC score | 0.995354 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS6_HUMAN
|
||||||
| NC score | 0.991056 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS11_HUMAN
|
||||||
| NC score | 0.963996 (rank : 5) | θ value | 2.47191e-82 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS9_HUMAN
|
||||||
| NC score | 0.956928 (rank : 6) | θ value | 8.77142e-88 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| NC score | 0.956417 (rank : 7) | θ value | 5.6855e-87 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS20_HUMAN
|
||||||
| NC score | 0.856927 (rank : 8) | θ value | 1.16704e-23 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS20_MOUSE
|
||||||
| NC score | 0.855878 (rank : 9) | θ value | 2.20094e-22 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS17_HUMAN
|
||||||
| NC score | 0.855553 (rank : 10) | θ value | 1.6852e-22 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS1_MOUSE
|
||||||
| NC score | 0.854807 (rank : 11) | θ value | 5.42112e-21 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS19_HUMAN
|
||||||
| NC score | 0.854721 (rank : 12) | θ value | 8.36355e-22 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS5_MOUSE
|
||||||
| NC score | 0.854676 (rank : 13) | θ value | 1.6852e-22 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS1_HUMAN
|
||||||
| NC score | 0.854554 (rank : 14) | θ value | 1.09232e-21 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS19_MOUSE
|
||||||
| NC score | 0.854066 (rank : 15) | θ value | 1.09232e-21 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS17_MOUSE
|
||||||
| NC score | 0.853260 (rank : 16) | θ value | 1.09232e-21 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS10_MOUSE
|
||||||
| NC score | 0.850592 (rank : 17) | θ value | 1.63225e-17 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS10_HUMAN
|
||||||
| NC score | 0.849691 (rank : 18) | θ value | 8.10077e-17 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS5_HUMAN
|
||||||
| NC score | 0.848032 (rank : 19) | θ value | 1.09232e-21 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS18_HUMAN
|
||||||
| NC score | 0.847210 (rank : 20) | θ value | 4.15078e-21 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS8_HUMAN
|
||||||
| NC score | 0.844342 (rank : 21) | θ value | 1.09232e-21 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS8_MOUSE
|
||||||
| NC score | 0.843314 (rank : 22) | θ value | 2.43343e-21 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS18_MOUSE
|
||||||
| NC score | 0.840938 (rank : 23) | θ value | 6.62687e-19 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS13_HUMAN
|
||||||
| NC score | 0.840586 (rank : 24) | θ value | 7.32683e-18 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS4_MOUSE
|
||||||
| NC score | 0.839357 (rank : 25) | θ value | 6.62687e-19 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS2_HUMAN
|
||||||
| NC score | 0.838651 (rank : 26) | θ value | 1.2077e-20 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS16_HUMAN
|
||||||
| NC score | 0.838064 (rank : 27) | θ value | 2.27762e-19 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS2_MOUSE
|
||||||
| NC score | 0.837463 (rank : 28) | θ value | 1.2077e-20 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS4_HUMAN
|
||||||
| NC score | 0.837204 (rank : 29) | θ value | 3.88503e-19 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS13_MOUSE
|
||||||
| NC score | 0.836049 (rank : 30) | θ value | 2.7842e-17 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS16_MOUSE
|
||||||
| NC score | 0.832421 (rank : 31) | θ value | 2.13179e-17 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS14_MOUSE
|
||||||
| NC score | 0.797139 (rank : 32) | θ value | 9.56915e-18 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS14_HUMAN
|
||||||
| NC score | 0.796584 (rank : 33) | θ value | 1.47631e-18 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.717752 (rank : 34) | θ value | 1.02238e-19 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.595529 (rank : 35) | θ value | 7.0802e-21 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.531297 (rank : 36) | θ value | 5.42112e-21 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
AXN1_MOUSE
|
||||||
| NC score | 0.509671 (rank : 37) | θ value | 0.00298849 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.497885 (rank : 38) | θ value | 0.00298849 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.493758 (rank : 39) | θ value | 0.000786445 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN1_HUMAN
|
||||||
| NC score | 0.491638 (rank : 40) | θ value | 0.0431538 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
SNX13_MOUSE
|
||||||
| NC score | 0.285252 (rank : 41) | θ value | 0.00665767 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
SNX13_HUMAN
|
||||||
| NC score | 0.284449 (rank : 42) | θ value | 0.00390308 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SNX25_HUMAN
|
||||||
| NC score | 0.243531 (rank : 43) | θ value | 8.40245e-06 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
AKA10_HUMAN
|
||||||
| NC score | 0.197709 (rank : 44) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| NC score | 0.197265 (rank : 45) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
GBG2_HUMAN
|
||||||
| NC score | 0.089599 (rank : 46) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59768, Q6P9A9 | Gene names | GNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit precursor (G gamma-I). | |||||
|
GBG2_MOUSE
|
||||||
| NC score | 0.089599 (rank : 47) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63213, P16874, Q3TYE8, Q61013, Q9TS47 | Gene names | Gng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-2 subunit precursor (G gamma-I). | |||||
|
GBG4_MOUSE
|
||||||
| NC score | 0.086164 (rank : 48) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50153 | Gene names | Gng4, Gngt4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-4 subunit precursor. | |||||
|
GBG4_HUMAN
|
||||||
| NC score | 0.081453 (rank : 49) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P50150 | Gene names | GNG4, GNGT4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-4 subunit precursor. | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.079306 (rank : 50) | θ value | 0.00869519 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
GBG8_HUMAN
|
||||||
| NC score | 0.079036 (rank : 51) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UK08 | Gene names | GNG8, GNG9, GNGT9 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-8 subunit precursor (Gamma-9). | |||||
|
GBG8_MOUSE
|
||||||
| NC score | 0.078994 (rank : 52) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63078, P43426 | Gene names | Gng8, Gng9, Gngt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-8 subunit precursor (Gamma-9). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.077409 (rank : 53) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.074941 (rank : 54) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
GBG7_HUMAN
|
||||||
| NC score | 0.074863 (rank : 55) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60262 | Gene names | GNG7, GNGT7 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-7 subunit precursor. | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.074307 (rank : 56) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.074244 (rank : 57) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
GBG3_HUMAN
|
||||||
| NC score | 0.070581 (rank : 58) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63215, P29798, Q61014 | Gene names | GNG3, GNGT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-3 subunit precursor. | |||||
|
GBG3_MOUSE
|
||||||
| NC score | 0.070581 (rank : 59) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63216, P29798, Q61014 | Gene names | Gng3, Gngt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-3 subunit precursor. | |||||
|
SNX14_MOUSE
|
||||||
| NC score | 0.064207 (rank : 60) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
GBG12_HUMAN
|
||||||
| NC score | 0.061023 (rank : 61) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UBI6, Q9BRV5 | Gene names | GNG12 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-12 subunit precursor. | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.060454 (rank : 62) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
GBG12_MOUSE
|
||||||
| NC score | 0.059165 (rank : 63) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DAS9, Q544P3 | Gene names | Gng12 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-12 subunit precursor. | |||||
|
GBG7_MOUSE
|
||||||
| NC score | 0.059142 (rank : 64) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61016, Q8JZP6 | Gene names | Gng7, Gngt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-7 subunit precursor. | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.054152 (rank : 65) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
SNX14_HUMAN
|
||||||
| NC score | 0.053786 (rank : 66) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.053259 (rank : 67) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
PREX1_HUMAN
|
||||||
| NC score | 0.051004 (rank : 68) | θ value | 0.0252991 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
SYEP_MOUSE
|
||||||
| NC score | 0.029874 (rank : 69) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGC7 | Gene names | Eprs, Qprs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional aminoacyl-tRNA synthetase [Includes: Glutamyl-tRNA synthetase (EC 6.1.1.17) (Glutamate--tRNA ligase); Prolyl-tRNA synthetase (EC 6.1.1.15) (Proline--tRNA ligase)]. | |||||
|
GAMT_MOUSE
|
||||||
| NC score | 0.017274 (rank : 70) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35969, Q3TZ58, Q3US90 | Gene names | Gamt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanidinoacetate N-methyltransferase (EC 2.1.1.2). | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.016539 (rank : 71) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.016425 (rank : 72) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ABLM3_HUMAN
|
||||||
| NC score | 0.014137 (rank : 73) | θ value | 0.21417 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94929, Q658S1, Q68CI5, Q9BV32 | Gene names | ABLIM3, KIAA0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.013692 (rank : 74) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
SMRD1_HUMAN
|
||||||
| NC score | 0.008508 (rank : 75) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96GM5, Q92924, Q9Y635 | Gene names | SMARCD1, BAF60A | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A). | |||||
|
SMRD1_MOUSE
|
||||||
| NC score | 0.008508 (rank : 76) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61466, P70384, Q8R0I7 | Gene names | Smarcd1, Baf60a, D15Kz1 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A) (D15KZ1 protein). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.008035 (rank : 77) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.002130 (rank : 78) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.002079 (rank : 79) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.000161 (rank : 80) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
RK_HUMAN
|
||||||
| NC score | -0.002173 (rank : 81) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15835 | Gene names | GRK1, RHOK | |||
|
Domain Architecture |

|
|||||
| Description | Rhodopsin kinase precursor (EC 2.7.11.14) (RK) (G protein-coupled receptor kinase 1). | |||||