Please be patient as the page loads
|
ZC3H5_MOUSE
|
||||||
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZC3H5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.982589 (rank : 2) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC3H5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
TTP_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 3) | NC score | 0.323892 (rank : 3) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 4) | NC score | 0.286738 (rank : 9) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 5) | NC score | 0.298382 (rank : 4) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
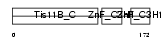
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISB_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 6) | NC score | 0.297757 (rank : 5) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
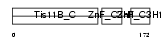
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 7) | NC score | 0.196939 (rank : 11) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
TISD_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 8) | NC score | 0.288813 (rank : 8) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |
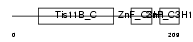
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TISD_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 9) | NC score | 0.294445 (rank : 7) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |
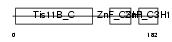
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TTP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 10) | NC score | 0.295885 (rank : 6) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
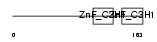
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
RC3H1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 11) | NC score | 0.195753 (rank : 12) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
MNAB_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 12) | NC score | 0.191029 (rank : 15) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
MNAB_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.191602 (rank : 14) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.037566 (rank : 96) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 15) | NC score | 0.263293 (rank : 10) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
RNF26_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.193913 (rank : 13) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BY78 | Gene names | RNF26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 26. | |||||
|
NIN_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.075639 (rank : 45) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.069682 (rank : 51) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.062701 (rank : 56) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.178773 (rank : 16) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.057582 (rank : 62) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.076665 (rank : 44) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.076769 (rank : 43) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
PML_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.074744 (rank : 47) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.168176 (rank : 17) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.065469 (rank : 55) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.051873 (rank : 67) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.032083 (rank : 101) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MGRN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.120825 (rank : 24) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D074, Q3U5V9, Q3UDA1, Q6ZQ97, Q8BZM9 | Gene names | Mgrn1, Kiaa0544 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1). | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.138825 (rank : 19) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.166530 (rank : 18) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.029767 (rank : 104) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
MGRN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.115372 (rank : 25) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.135056 (rank : 22) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.136817 (rank : 20) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
PAX9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.037772 (rank : 95) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |
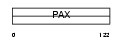
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
ZN217_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.016278 (rank : 120) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75362 | Gene names | ZNF217, ZABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 217. | |||||
|
BIRC4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.087473 (rank : 36) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |
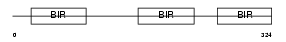
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.059206 (rank : 60) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
MIB2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.042321 (rank : 85) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
PRC1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.079530 (rank : 40) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
CGRF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.110754 (rank : 27) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99675, Q96BX2 | Gene names | CGRRF1, CGR19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
CT012_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.039225 (rank : 89) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C008, Q8C060 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C20orf12 homolog. | |||||
|
HINT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.099783 (rank : 30) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49773, Q9H5W8 | Gene names | HINT1, HINT, PKCI1, PRKCNH1 | |||
|
Domain Architecture |
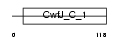
|
|||||
| Description | Histidine triad nucleotide-binding protein 1 (Adenosine 5'- monophosphoramidase) (Protein kinase C inhibitor 1) (Protein kinase C- interacting protein 1) (PKCI-1). | |||||
|
HINT1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.100132 (rank : 29) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70349 | Gene names | Hint1, Hint, Pkci, Pkci1, Prkcnh1 | |||
|
Domain Architecture |
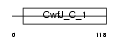
|
|||||
| Description | Histidine triad nucleotide-binding protein 1 (Adenosine 5'- monophosphoramidase) (Protein kinase C inhibitor 1) (Protein kinase C- interacting protein 1) (PKCI-1). | |||||
|
MIB2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.041276 (rank : 88) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.131004 (rank : 23) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.047555 (rank : 76) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SUHW3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.011268 (rank : 131) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
TLK2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.014012 (rank : 124) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
BIRC4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.083648 (rank : 38) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |
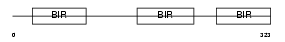
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.097035 (rank : 31) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.060147 (rank : 59) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.053149 (rank : 66) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
RXRA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.010094 (rank : 133) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |
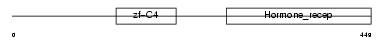
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
TAF12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.077640 (rank : 42) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16514, Q15775 | Gene names | TAF12, TAF15, TAF2J, TAFII20 | |||
|
Domain Architecture |
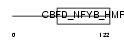
|
|||||
| Description | Transcription initiation factor TFIID subunit 12 (Transcription initiation factor TFIID 20/15 kDa subunits) (TAFII-20/TAFII-15) (TAFII20/TAFII15). | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.069282 (rank : 53) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
CGRF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.115216 (rank : 26) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BMJ7, Q3U3F1, Q8CI24 | Gene names | Cgrrf1, Cgr19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.062414 (rank : 57) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.060273 (rank : 58) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
BIRC8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.089448 (rank : 35) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96P09, Q96RW5 | Gene names | BIRC8, ILP2 | |||
|
Domain Architecture |
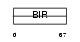
|
|||||
| Description | Baculoviral IAP repeat-containing protein 8 (Inhibitor of apoptosis- like protein 2) (IAP-like protein 2) (ILP-2) (Testis-specific inhibitor of apoptosis). | |||||
|
HOMEZ_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.053218 (rank : 65) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W88 | Gene names | Homez, Kiaa1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
PIDD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.013556 (rank : 126) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERV7 | Gene names | Lrdd, Pidd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and death domain-containing protein. | |||||
|
PKNX2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.019426 (rank : 114) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BG99, Q8R4B0 | Gene names | Pknox2, Prep2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein PKNOX2 (PBX/knotted homeobox 2) (Homeobox protein PREP-2). | |||||
|
R113A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.094681 (rank : 34) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
R113B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.095418 (rank : 33) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.135736 (rank : 21) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.051699 (rank : 68) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.026690 (rank : 106) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.050181 (rank : 71) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.026580 (rank : 107) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.056517 (rank : 64) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PMYT1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.006344 (rank : 137) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESG9, Q8R3L4 | Gene names | Pkmyt1, Myt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase (EC 2.7.11.1) (Myt1 kinase). | |||||
|
TRM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.045421 (rank : 78) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
BIRC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.075010 (rank : 46) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
FLNB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.015190 (rank : 122) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
K2C1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.024096 (rank : 111) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7Z794, Q7RTS8 | Gene names | KRT1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b. | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.103032 (rank : 28) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
BIRC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.073119 (rank : 48) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.010748 (rank : 132) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.038786 (rank : 91) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
K0562_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.025812 (rank : 108) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60308, Q5JSQ3, Q5SR24, Q5SR25, Q6PKF5, Q86W32, Q86X14 | Gene names | KIAA0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
MEOX2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.009366 (rank : 134) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32443, Q544T6 | Gene names | Meox2, Gax, Mox-2, Mox2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.096266 (rank : 32) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.038629 (rank : 92) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.039163 (rank : 90) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.047777 (rank : 75) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.048144 (rank : 74) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MEOX2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.008787 (rank : 136) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50222, O75263, Q9UPL6 | Gene names | MEOX2, GAX, MOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2) (Growth arrest-specific homeobox). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.035342 (rank : 98) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.038287 (rank : 93) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
ZFP95_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | -0.000540 (rank : 140) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |
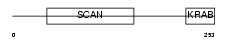
|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||
|
BIRC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.069306 (rank : 52) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.069995 (rank : 49) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BRCA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.049817 (rank : 72) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
CENPQ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.035374 (rank : 97) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CPQ5, Q8C2G4, Q8VEF8, Q9CX70 | Gene names | Cenpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
DUS3L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.069720 (rank : 50) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.027201 (rank : 105) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.005392 (rank : 138) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
JIP3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.033109 (rank : 100) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.018628 (rank : 118) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.037942 (rank : 94) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.012372 (rank : 130) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.043019 (rank : 83) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.043026 (rank : 82) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.024982 (rank : 110) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCEA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.013389 (rank : 127) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15560, Q8TD37, Q8TD38 | Gene names | TCEA2 | |||
|
Domain Architecture |
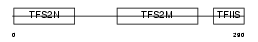
|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Transcription elongation factor TFIIS.l). | |||||
|
TRAF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.025062 (rank : 109) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00463 | Gene names | TRAF5, RNF84 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5 (RING finger protein 84). | |||||
|
U383_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.031185 (rank : 102) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2Q2, Q3V1K3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.015635 (rank : 121) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CEP41_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.019068 (rank : 115) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.043495 (rank : 81) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.051077 (rank : 70) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.045618 (rank : 77) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.044712 (rank : 79) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.017505 (rank : 119) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
RABE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.043507 (rank : 80) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |
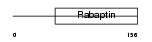
|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.051110 (rank : 69) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.042911 (rank : 84) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TUFT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.049727 (rank : 73) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
ARRD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.012550 (rank : 129) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96B67, Q9P2H1 | Gene names | ARRDC3, KIAA1376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 3. | |||||
|
ARRD3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.012708 (rank : 128) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPQ9, Q4KMN0, Q80TE6, Q8K0L7 | Gene names | Arrdc3, Kiaa1376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 3. | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.021957 (rank : 112) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
CASC5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.014124 (rank : 123) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
CLPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.020890 (rank : 113) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H078, Q8ND11, Q9H8Y0 | Gene names | CLPB, SKD3 | |||
|
Domain Architecture |

|
|||||
| Description | Caseinolytic peptidase B protein homolog (Suppressor of potassium transport defect 3). | |||||
|
F13B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.009325 (rank : 135) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.041504 (rank : 87) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.030859 (rank : 103) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.033161 (rank : 99) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.068768 (rank : 54) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
NEU1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.013736 (rank : 125) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01178, Q3MIG0 | Gene names | OXT, OT | |||
|
Domain Architecture |
|
|||||
| Description | Oxytocin-neurophysin 1 precursor (OT-NPI) [Contains: Oxytocin (Ocytocin); Neurophysin 1]. | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.004060 (rank : 139) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.041670 (rank : 86) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.057791 (rank : 61) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
U2AF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.018960 (rank : 116) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01081 | Gene names | U2AF1, U2AF35 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
U2AF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.018951 (rank : 117) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D883, Q564E4, Q99LX2, Q9CZ98 | Gene names | U2af1 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
BIRC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.057202 (rank : 63) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96CA5, Q9BQV0, Q9H2A8, Q9HAP7 | Gene names | BIRC7, KIAP, LIVIN, MLIAP | |||
|
Domain Architecture |
|
|||||
| Description | Baculoviral IAP repeat-containing protein 7 (Kidney inhibitor of apoptosis protein) (KIAP) (Melanoma inhibitor of apoptosis protein) (ML-IAP) (Livin). | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.079230 (rank : 41) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.084253 (rank : 37) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.082142 (rank : 39) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC3H5_HUMAN
|
||||||
| NC score | 0.982589 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.323892 (rank : 3) | θ value | 7.1131e-05 (rank : 3) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.298382 (rank : 4) | θ value | 0.00102713 (rank : 5) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.297757 (rank : 5) | θ value | 0.00134147 (rank : 6) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.295885 (rank : 6) | θ value | 0.00298849 (rank : 10) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TISD_MOUSE
|
||||||
| NC score | 0.294445 (rank : 7) | θ value | 0.00298849 (rank : 9) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TISD_HUMAN
|
||||||
| NC score | 0.288813 (rank : 8) | θ value | 0.00298849 (rank : 8) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.286738 (rank : 9) | θ value | 9.29e-05 (rank : 4) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.263293 (rank : 10) | θ value | 0.0113563 (rank : 15) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.196939 (rank : 11) | θ value | 0.00228821 (rank : 7) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
RC3H1_MOUSE
|
||||||
| NC score | 0.195753 (rank : 12) | θ value | 0.00665767 (rank : 11) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q4VGL6, Q69Z31 | Gene names | Rc3h1, Gm551, Kiaa2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1) (Sanroque protein). | |||||
|
RNF26_HUMAN
|
||||||
| NC score | 0.193913 (rank : 13) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BY78 | Gene names | RNF26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 26. | |||||
|
MNAB_MOUSE
|
||||||
| NC score | 0.191602 (rank : 14) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
MNAB_HUMAN
|
||||||
| NC score | 0.191029 (rank : 15) | θ value | 0.00869519 (rank : 12) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HBD1, Q5JPD7, Q86ST6, Q8N3D6, Q96F27, Q9H5J2, Q9HBD2, Q9NWN9, Q9NXE1 | Gene names | MNAB, RNF164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein (RING finger protein 164). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.178773 (rank : 16) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.168176 (rank : 17) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.166530 (rank : 18) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.138825 (rank : 19) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.136817 (rank : 20) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.135736 (rank : 21) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.135056 (rank : 22) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.131004 (rank : 23) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MGRN1_MOUSE
|
||||||
| NC score | 0.120825 (rank : 24) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D074, Q3U5V9, Q3UDA1, Q6ZQ97, Q8BZM9 | Gene names | Mgrn1, Kiaa0544 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1). | |||||
|
MGRN1_HUMAN
|
||||||
| NC score | 0.115372 (rank : 25) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
CGRF1_MOUSE
|
||||||
| NC score | 0.115216 (rank : 26) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BMJ7, Q3U3F1, Q8CI24 | Gene names | Cgrrf1, Cgr19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
CGRF1_HUMAN
|
||||||
| NC score | 0.110754 (rank : 27) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99675, Q96BX2 | Gene names | CGRRF1, CGR19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with RING finger domain 1 (Cell growth regulatory gene 19 protein). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.103032 (rank : 28) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
HINT1_MOUSE
|
||||||
| NC score | 0.100132 (rank : 29) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70349 | Gene names | Hint1, Hint, Pkci, Pkci1, Prkcnh1 | |||
|
Domain Architecture |

|
|||||
| Description | Histidine triad nucleotide-binding protein 1 (Adenosine 5'- monophosphoramidase) (Protein kinase C inhibitor 1) (Protein kinase C- interacting protein 1) (PKCI-1). | |||||
|
HINT1_HUMAN
|
||||||
| NC score | 0.099783 (rank : 30) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49773, Q9H5W8 | Gene names | HINT1, HINT, PKCI1, PRKCNH1 | |||
|
Domain Architecture |

|
|||||
| Description | Histidine triad nucleotide-binding protein 1 (Adenosine 5'- monophosphoramidase) (Protein kinase C inhibitor 1) (Protein kinase C- interacting protein 1) (PKCI-1). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.097035 (rank : 31) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.096266 (rank : 32) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
R113B_HUMAN
|
||||||
| NC score | 0.095418 (rank : 33) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
R113A_HUMAN
|
||||||
| NC score | 0.094681 (rank : 34) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
BIRC8_HUMAN
|
||||||
| NC score | 0.089448 (rank : 35) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96P09, Q96RW5 | Gene names | BIRC8, ILP2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 8 (Inhibitor of apoptosis- like protein 2) (IAP-like protein 2) (ILP-2) (Testis-specific inhibitor of apoptosis). | |||||
|
BIRC4_HUMAN
|
||||||
| NC score | 0.087473 (rank : 36) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.084253 (rank : 37) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
BIRC4_MOUSE
|
||||||
| NC score | 0.083648 (rank : 38) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.082142 (rank : 39) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
PRC1_MOUSE
|
||||||
| NC score | 0.079530 (rank : 40) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.079230 (rank : 41) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
TAF12_HUMAN
|
||||||
| NC score | 0.077640 (rank : 42) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16514, Q15775 | Gene names | TAF12, TAF15, TAF2J, TAFII20 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 12 (Transcription initiation factor TFIID 20/15 kDa subunits) (TAFII-20/TAFII-15) (TAFII20/TAFII15). | |||||
|
MYLIP_HUMAN
|
||||||
| NC score | 0.076769 (rank : 43) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WY64, Q9BU73, Q9NRL9, Q9UHE7 | Gene names | MYLIP, BZF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.076665 (rank : 44) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.075639 (rank : 45) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
BIRC2_HUMAN
|
||||||
| NC score | 0.075010 (rank : 46) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
PML_HUMAN
|
||||||
| NC score | 0.074744 (rank : 47) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29590, P29591, P29592, P29593, Q00755 | Gene names | PML, MYL, RNF71, TRIM19 | |||
|
Domain Architecture |

|
|||||
| Description | Probable transcription factor PML (Tripartite motif-containing protein 19) (RING finger protein 71). | |||||
|
BIRC2_MOUSE
|
||||||
| NC score | 0.073119 (rank : 48) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC3_MOUSE
|
||||||
| NC score | 0.069995 (rank : 49) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
DUS3L_HUMAN
|
||||||
| NC score | 0.069720 (rank : 50) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.069682 (rank : 51) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BIRC3_HUMAN
|
||||||
| NC score | 0.069306 (rank : 52) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.069282 (rank : 53) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.068768 (rank : 54) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.065469 (rank : 55) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.062701 (rank : 56) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.062414 (rank : 57) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.060273 (rank : 58) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.060147 (rank : 59) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.059206 (rank : 60) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.057791 (rank : 61) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.057582 (rank : 62) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
BIRC7_HUMAN
|
||||||
| NC score | 0.057202 (rank : 63) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96CA5, Q9BQV0, Q9H2A8, Q9HAP7 | Gene names | BIRC7, KIAP, LIVIN, MLIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 7 (Kidney inhibitor of apoptosis protein) (KIAP) (Melanoma inhibitor of apoptosis protein) (ML-IAP) (Livin). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.056517 (rank : 64) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
HOMEZ_MOUSE
|
||||||
| NC score | 0.053218 (rank : 65) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W88 | Gene names | Homez, Kiaa1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.053149 (rank : 66) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.051873 (rank : 67) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.051699 (rank : 68) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.051110 (rank : 69) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.051077 (rank : 70) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.050181 (rank : 71) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
BRCA2_MOUSE
|
||||||
| NC score | 0.049817 (rank : 72) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
TUFT1_MOUSE
|
||||||
| NC score | 0.049727 (rank : 73) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08970 | Gene names | Tuft1 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.048144 (rank : 74) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.047777 (rank : 75) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.047555 (rank : 76) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.045618 (rank : 77) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
TRM1_HUMAN
|
||||||
| NC score | 0.045421 (rank : 78) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.044712 (rank : 79) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
RABE2_HUMAN
|
||||||
| NC score | 0.043507 (rank : 80) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.043495 (rank : 81) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.043026 (rank : 82) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.043019 (rank : 83) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.042911 (rank : 84) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.042321 (rank : 85) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.041670 (rank : 86) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.041504 (rank : 87) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
MIB2_MOUSE
|
||||||
| NC score | 0.041276 (rank : 88) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
CT012_MOUSE
|
||||||
| NC score | 0.039225 (rank : 89) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C008, Q8C060 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C20orf12 homolog. | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.039163 (rank : 90) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.038786 (rank : 91) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.038629 (rank : 92) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.038287 (rank : 93) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.037942 (rank : 94) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PAX9_HUMAN
|
||||||
| NC score | 0.037772 (rank : 95) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55771, Q99582, Q9UQR4 | Gene names | PAX9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.037566 (rank : 96) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
CENPQ_MOUSE
|
||||||
| NC score | 0.035374 (rank : 97) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CPQ5, Q8C2G4, Q8VEF8, Q9CX70 | Gene names | Cenpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.035342 (rank : 98) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.033161 (rank : 99) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
JIP3_MOUSE
|
||||||
| NC score | 0.033109 (rank : 100) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.032083 (rank : 101) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
U383_MOUSE
|
||||||
| NC score | 0.031185 (rank : 102) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2Q2, Q3V1K3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.030859 (rank : 103) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.029767 (rank : 104) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.027201 (rank : 105) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.026690 (rank : 106) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.026580 (rank : 107) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
K0562_HUMAN
|
||||||
| NC score | 0.025812 (rank : 108) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60308, Q5JSQ3, Q5SR24, Q5SR25, Q6PKF5, Q86W32, Q86X14 | Gene names | KIAA0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
TRAF5_HUMAN
|
||||||
| NC score | 0.025062 (rank : 109) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00463 | Gene names | TRAF5, RNF84 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5 (RING finger protein 84). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.024982 (rank : 110) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
K2C1B_HUMAN
|
||||||
| NC score | 0.024096 (rank : 111) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7Z794, Q7RTS8 | Gene names | KRT1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b. | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.021957 (rank : 112) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
CLPB_HUMAN
|
||||||
| NC score | 0.020890 (rank : 113) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H078, Q8ND11, Q9H8Y0 | Gene names | CLPB, SKD3 | |||
|
Domain Architecture |

|
|||||
| Description | Caseinolytic peptidase B protein homolog (Suppressor of potassium transport defect 3). | |||||
|
PKNX2_MOUSE
|
||||||
| NC score | 0.019426 (rank : 114) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BG99, Q8R4B0 | Gene names | Pknox2, Prep2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein PKNOX2 (PBX/knotted homeobox 2) (Homeobox protein PREP-2). | |||||
|
CEP41_HUMAN
|
||||||
| NC score | 0.019068 (rank : 115) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
U2AF1_HUMAN
|
||||||
| NC score | 0.018960 (rank : 116) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01081 | Gene names | U2AF1, U2AF35 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
U2AF1_MOUSE
|
||||||
| NC score | 0.018951 (rank : 117) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D883, Q564E4, Q99LX2, Q9CZ98 | Gene names | U2af1 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 35 kDa subunit (U2 auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary factor small subunit). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.018628 (rank : 118) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.017505 (rank : 119) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ZN217_HUMAN
|
||||||
| NC score | 0.016278 (rank : 120) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75362 | Gene names | ZNF217, ZABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 217. | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.015635 (rank : 121) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
FLNB_MOUSE
|
||||||
| NC score | 0.015190 (rank : 122) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80X90, Q8VHX4, Q8VHX7, Q99KY3 | Gene names | Flnb | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (ABP- 280-like protein). | |||||
|
CASC5_MOUSE
|
||||||
| NC score | 0.014124 (rank : 123) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q66JQ7, Q8CCH6, Q9CV38 | Gene names | Casc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein homolog). | |||||
|
TLK2_HUMAN
|
||||||
| NC score | 0.014012 (rank : 124) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
NEU1_HUMAN
|
||||||
| NC score | 0.013736 (rank : 125) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01178, Q3MIG0 | Gene names | OXT, OT | |||
|
Domain Architecture |

|
|||||
| Description | Oxytocin-neurophysin 1 precursor (OT-NPI) [Contains: Oxytocin (Ocytocin); Neurophysin 1]. | |||||
|
PIDD_MOUSE
|
||||||
| NC score | 0.013556 (rank : 126) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERV7 | Gene names | Lrdd, Pidd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat and death domain-containing protein. | |||||
|
TCEA2_HUMAN
|
||||||
| NC score | 0.013389 (rank : 127) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15560, Q8TD37, Q8TD38 | Gene names | TCEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Transcription elongation factor TFIIS.l). | |||||
|
ARRD3_MOUSE
|
||||||
| NC score | 0.012708 (rank : 128) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPQ9, Q4KMN0, Q80TE6, Q8K0L7 | Gene names | Arrdc3, Kiaa1376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 3. | |||||
|
ARRD3_HUMAN
|
||||||
| NC score | 0.012550 (rank : 129) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96B67, Q9P2H1 | Gene names | ARRDC3, KIAA1376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 3. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.012372 (rank : 130) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SUHW3_HUMAN
|
||||||
| NC score | 0.011268 (rank : 131) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.010748 (rank : 132) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
RXRA_HUMAN
|
||||||
| NC score | 0.010094 (rank : 133) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
MEOX2_MOUSE
|
||||||
| NC score | 0.009366 (rank : 134) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32443, Q544T6 | Gene names | Meox2, Gax, Mox-2, Mox2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2). | |||||
|
F13B_MOUSE
|
||||||
| NC score | 0.009325 (rank : 135) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07968 | Gene names | F13b, Cf13b | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII B chain precursor (Protein-glutamine gamma- glutamyltransferase B chain) (Transglutaminase B chain). | |||||
|
MEOX2_HUMAN
|
||||||
| NC score | 0.008787 (rank : 136) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50222, O75263, Q9UPL6 | Gene names | MEOX2, GAX, MOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-2 (Mesenchyme homeobox 2) (Growth arrest-specific homeobox). | |||||
|
PMYT1_MOUSE
|
||||||
| NC score | 0.006344 (rank : 137) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESG9, Q8R3L4 | Gene names | Pkmyt1, Myt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase (EC 2.7.11.1) (Myt1 kinase). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.005392 (rank : 138) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.004060 (rank : 139) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
ZFP95_HUMAN
|
||||||
| NC score | -0.000540 (rank : 140) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||