Please be patient as the page loads
|
DUS3L_HUMAN
|
||||||
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DUS3L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
DUS3L_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988280 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XI1, Q3TZQ1, Q7TT12 | Gene names | Dus3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
DUS1L_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 3) | NC score | 0.601279 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1R4, Q96AI3 | Gene names | DUS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 1-like (EC 1.-.-.-). | |||||
|
DUS1L_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 4) | NC score | 0.599230 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C2P3, Q8VCN7, Q9D124 | Gene names | Dus1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 1-like (EC 1.-.-.-). | |||||
|
DUS4L_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 5) | NC score | 0.601533 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95620, Q2NKK1 | Gene names | DUS4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 4-like (EC 1.-.-.-) (PP35). | |||||
|
DUS2L_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 6) | NC score | 0.563299 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D7B1, Q6PDX2 | Gene names | Dus2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 2-like (EC 1.-.-.-). | |||||
|
DUS4L_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 7) | NC score | 0.593386 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q32M08, Q9D6P4 | Gene names | Dus4l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 4-like (EC 1.-.-.-). | |||||
|
DUS2L_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 8) | NC score | 0.562490 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NX74, Q4H4D9 | Gene names | DUS2L, DUS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 2-like (EC 1.-.-.-) (hDUS2) (Up-regulated in lung cancer) (URLC8). | |||||
|
TISB_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 9) | NC score | 0.277824 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
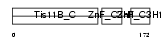
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 10) | NC score | 0.275990 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
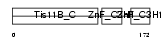
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISD_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 11) | NC score | 0.268760 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |
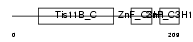
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TISD_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 12) | NC score | 0.273727 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |
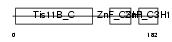
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TTP_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 13) | NC score | 0.266345 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
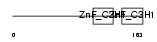
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.249127 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.079871 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.087625 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.093170 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
SAS10_MOUSE
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.039326 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI13, Q3UL95, Q8R5C6, Q9JJ12 | Gene names | Crlz1, Crl1, Sas10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1) (Crl-1). | |||||
|
SH3BG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.033428 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55822, Q9BRB8 | Gene names | SH3BGR | |||
|
Domain Architecture |
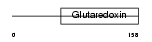
|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein) (21- glutamic acid-rich protein) (21-GARP). | |||||
|
ZC3H5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.073753 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.003419 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
STAG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.016555 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.005808 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.004957 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.003769 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.059054 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.010002 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.081472 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.007686 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
ZC3H5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.069720 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
AN32A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.012458 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.012458 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
BRD7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.010987 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
CASL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.009734 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
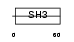
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.005151 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.067776 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.056556 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
PAR14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.010687 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.063570 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ERRFI_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.012748 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.061393 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.052270 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
SCPDH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.023237 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R127, Q3TMB9 | Gene names | Sccpdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable saccharopine dehydrogenase (EC 1.5.1.9). | |||||
|
ZHX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.007575 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.067777 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
CS007_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.067406 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.060250 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
DUS3L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
DUS3L_MOUSE
|
||||||
| NC score | 0.988280 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XI1, Q3TZQ1, Q7TT12 | Gene names | Dus3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
DUS4L_HUMAN
|
||||||
| NC score | 0.601533 (rank : 3) | θ value | 3.76295e-14 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95620, Q2NKK1 | Gene names | DUS4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 4-like (EC 1.-.-.-) (PP35). | |||||
|
DUS1L_HUMAN
|
||||||
| NC score | 0.601279 (rank : 4) | θ value | 2.35696e-16 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1R4, Q96AI3 | Gene names | DUS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 1-like (EC 1.-.-.-). | |||||
|
DUS1L_MOUSE
|
||||||
| NC score | 0.599230 (rank : 5) | θ value | 5.25075e-16 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C2P3, Q8VCN7, Q9D124 | Gene names | Dus1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 1-like (EC 1.-.-.-). | |||||
|
DUS4L_MOUSE
|
||||||
| NC score | 0.593386 (rank : 6) | θ value | 4.16044e-13 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q32M08, Q9D6P4 | Gene names | Dus4l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 4-like (EC 1.-.-.-). | |||||
|
DUS2L_MOUSE
|
||||||
| NC score | 0.563299 (rank : 7) | θ value | 1.86753e-13 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D7B1, Q6PDX2 | Gene names | Dus2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 2-like (EC 1.-.-.-). | |||||
|
DUS2L_HUMAN
|
||||||
| NC score | 0.562490 (rank : 8) | θ value | 7.09661e-13 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NX74, Q4H4D9 | Gene names | DUS2L, DUS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 2-like (EC 1.-.-.-) (hDUS2) (Up-regulated in lung cancer) (URLC8). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.277824 (rank : 9) | θ value | 0.000158464 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.275990 (rank : 10) | θ value | 0.00020696 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISD_MOUSE
|
||||||
| NC score | 0.273727 (rank : 11) | θ value | 0.00020696 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
TISD_HUMAN
|
||||||
| NC score | 0.268760 (rank : 12) | θ value | 0.00020696 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.266345 (rank : 13) | θ value | 0.000461057 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.249127 (rank : 14) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.093170 (rank : 15) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.087625 (rank : 16) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.081472 (rank : 17) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.079871 (rank : 18) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
ZC3H5_HUMAN
|
||||||
| NC score | 0.073753 (rank : 19) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC3H5_MOUSE
|
||||||
| NC score | 0.069720 (rank : 20) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.067777 (rank : 21) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.067776 (rank : 22) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.067406 (rank : 23) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.063570 (rank : 24) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.061393 (rank : 25) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.060250 (rank : 26) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.059054 (rank : 27) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.056556 (rank : 28) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.052270 (rank : 29) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
SAS10_MOUSE
|
||||||
| NC score | 0.039326 (rank : 30) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JI13, Q3UL95, Q8R5C6, Q9JJ12 | Gene names | Crlz1, Crl1, Sas10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Something about silencing protein 10 (Disrupter of silencing SAS10) (Charged amino acid-rich leucine zipper 1) (Crl-1). | |||||
|
SH3BG_HUMAN
|
||||||
| NC score | 0.033428 (rank : 31) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55822, Q9BRB8 | Gene names | SH3BGR | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein) (21- glutamic acid-rich protein) (21-GARP). | |||||
|
SCPDH_MOUSE
|
||||||
| NC score | 0.023237 (rank : 32) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R127, Q3TMB9 | Gene names | Sccpdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable saccharopine dehydrogenase (EC 1.5.1.9). | |||||
|
STAG1_HUMAN
|
||||||
| NC score | 0.016555 (rank : 33) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVM7, O00539 | Gene names | STAG1, SA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
ERRFI_MOUSE
|
||||||
| NC score | 0.012748 (rank : 34) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
AN32A_MOUSE
|
||||||
| NC score | 0.012458 (rank : 35) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| NC score | 0.012458 (rank : 36) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.010987 (rank : 37) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
PAR14_MOUSE
|
||||||
| NC score | 0.010687 (rank : 38) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.010002 (rank : 39) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CASL_HUMAN
|
||||||
| NC score | 0.009734 (rank : 40) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.007686 (rank : 41) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
ZHX1_MOUSE
|
||||||
| NC score | 0.007575 (rank : 42) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.005808 (rank : 43) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.005151 (rank : 44) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.004957 (rank : 45) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.003769 (rank : 46) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.003419 (rank : 47) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||