Please be patient as the page loads
|
BRD7_HUMAN
|
||||||
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BRD7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.982216 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD9_MOUSE
|
||||||
| θ value | 5.34618e-69 (rank : 3) | NC score | 0.920355 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
BRD9_HUMAN
|
||||||
| θ value | 3.83135e-67 (rank : 4) | NC score | 0.921963 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 5) | NC score | 0.496655 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 6) | NC score | 0.471312 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 7) | NC score | 0.496036 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 8) | NC score | 0.565822 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 9) | NC score | 0.531396 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 10) | NC score | 0.557341 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
PCAF_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 11) | NC score | 0.550258 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 12) | NC score | 0.425358 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 13) | NC score | 0.523692 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 14) | NC score | 0.535941 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 15) | NC score | 0.534097 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 16) | NC score | 0.535102 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 17) | NC score | 0.443612 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 18) | NC score | 0.437893 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TAF1L_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 19) | NC score | 0.517686 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 20) | NC score | 0.486614 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 21) | NC score | 0.481485 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BRWD1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 22) | NC score | 0.287447 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 23) | NC score | 0.268944 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 24) | NC score | 0.427760 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 25) | NC score | 0.496759 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 26) | NC score | 0.214540 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 27) | NC score | 0.511059 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 28) | NC score | 0.420568 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 29) | NC score | 0.426572 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 30) | NC score | 0.419672 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 31) | NC score | 0.213950 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 32) | NC score | 0.419352 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 33) | NC score | 0.425382 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 34) | NC score | 0.262291 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 35) | NC score | 0.392472 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 36) | NC score | 0.278041 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
BRDT_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 37) | NC score | 0.510295 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 38) | NC score | 0.357293 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 39) | NC score | 0.485360 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 40) | NC score | 0.478471 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 41) | NC score | 0.360067 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 42) | NC score | 0.287085 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.307923 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 44) | NC score | 0.280659 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 45) | NC score | 0.301695 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
PCDG2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 46) | NC score | 0.020512 (rank : 123) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5H1, Q9Y5D5 | Gene names | PCDHGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A2 precursor (PCDH-gamma-A2). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.304579 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TTF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.065021 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.045856 (rank : 113) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.202425 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.204374 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
CF188_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.048294 (rank : 112) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R100, Q8C3G8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf188 homolog. | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.008092 (rank : 148) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.025664 (rank : 118) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.062409 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
WDR63_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.055037 (rank : 102) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWG1, Q96L72, Q96NU4 | Gene names | WDR63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 63 (Testis development protein NYD-SP29). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.019550 (rank : 124) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
PCDGC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.015211 (rank : 131) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.023650 (rank : 121) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.009374 (rank : 143) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
MDM2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.023693 (rank : 120) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23804, Q61040, Q64330 | Gene names | Mdm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.065352 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.041130 (rank : 114) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PCDG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.014835 (rank : 133) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5H4, Q9Y5D6 | Gene names | PCDHGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A1 precursor (PCDH-gamma-A1). | |||||
|
RXRA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.014900 (rank : 132) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28700 | Gene names | Rxra, Nr2b1 | |||
|
Domain Architecture |
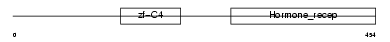
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
PCDGJ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.013461 (rank : 137) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5F8, Q9UN63 | Gene names | PCDHGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B7 precursor (PCDH-gamma-B7). | |||||
|
CM003_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.030114 (rank : 117) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C263 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3 homolog. | |||||
|
K0408_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.025402 (rank : 119) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZU52, O43158, Q5TF20, Q7L2M2 | Gene names | KIAA0408 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0408. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.018959 (rank : 127) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.015968 (rank : 130) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
PCDG7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.014138 (rank : 135) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.032378 (rank : 115) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
RXRA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.014208 (rank : 134) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |
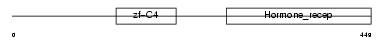
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.012695 (rank : 138) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
HBS1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.009435 (rank : 142) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
SEPT4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.009301 (rank : 145) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28661, Q5ND15 | Gene names | Sept4, Pnutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-4 (Peanut-like protein 2) (Brain protein H5). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.018593 (rank : 128) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
TBCD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.008477 (rank : 147) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.005299 (rank : 151) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
DUS3L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.010987 (rank : 140) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.010999 (rank : 139) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.009335 (rank : 144) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
ITF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.018993 (rank : 126) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.016497 (rank : 129) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
PCDG3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.013802 (rank : 136) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5H0, Q9Y5D4 | Gene names | PCDHGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A3 precursor (PCDH-gamma-A3). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.010880 (rank : 141) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.019496 (rank : 125) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
CC14B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.006333 (rank : 149) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.009023 (rank : 146) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CF188_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.021891 (rank : 122) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5C1 | Gene names | C6orf188 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf188. | |||||
|
LMNB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.002604 (rank : 152) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.030151 (rank : 116) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.005618 (rank : 150) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | -0.003825 (rank : 153) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
AF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.094187 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.096360 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.105816 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.113053 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.111601 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.077944 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.074402 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.074953 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.075264 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.060947 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.067108 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.055680 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.052413 (rank : 108) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JADE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.106656 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.106639 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.103421 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.101616 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.107397 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.111138 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JHD3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.054256 (rank : 106) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054966 (rank : 103) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.056993 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.055498 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.053174 (rank : 107) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.054942 (rank : 104) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
LY10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.125549 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.057884 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.067961 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.067304 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MTF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.051607 (rank : 109) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |
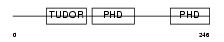
|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.051197 (rank : 110) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.071554 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.064163 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.066067 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.062483 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.062507 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.067032 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.099703 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.102125 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.071328 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.069670 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.081889 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.080537 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.086858 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.086221 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.102554 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.101865 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.058786 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.058512 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
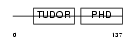
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.073063 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.070538 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.106231 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SP110_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.058060 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.103302 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.101207 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.055338 (rank : 101) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.057617 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050488 (rank : 111) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.054421 (rank : 105) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| NC score | 0.982216 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD9_HUMAN
|
||||||
| NC score | 0.921963 (rank : 3) | θ value | 3.83135e-67 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD9_MOUSE
|
||||||
| NC score | 0.920355 (rank : 4) | θ value | 5.34618e-69 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.565822 (rank : 5) | θ value | 9.59137e-10 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.557341 (rank : 6) | θ value | 4.76016e-09 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
PCAF_HUMAN
|
||||||
| NC score | 0.550258 (rank : 7) | θ value | 1.06045e-08 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.535941 (rank : 8) | θ value | 2.36244e-08 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.535102 (rank : 9) | θ value | 2.36244e-08 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.534097 (rank : 10) | θ value | 2.36244e-08 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.531396 (rank : 11) | θ value | 3.64472e-09 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.523692 (rank : 12) | θ value | 1.80886e-08 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
TAF1L_HUMAN
|
||||||
| NC score | 0.517686 (rank : 13) | θ value | 1.99992e-07 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.511059 (rank : 14) | θ value | 3.77169e-06 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
BRDT_MOUSE
|
||||||
| NC score | 0.510295 (rank : 15) | θ value | 7.1131e-05 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.496759 (rank : 16) | θ value | 2.21117e-06 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.496655 (rank : 17) | θ value | 6.20254e-17 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.496036 (rank : 18) | θ value | 6.85773e-16 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.486614 (rank : 19) | θ value | 3.41135e-07 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.485360 (rank : 20) | θ value | 0.000158464 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.481485 (rank : 21) | θ value | 3.41135e-07 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.478471 (rank : 22) | θ value | 0.000270298 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.471312 (rank : 23) | θ value | 6.20254e-17 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.443612 (rank : 24) | θ value | 3.08544e-08 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.437893 (rank : 25) | θ value | 5.26297e-08 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.427760 (rank : 26) | θ value | 2.21117e-06 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.426572 (rank : 27) | θ value | 4.92598e-06 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.425382 (rank : 28) | θ value | 1.09739e-05 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.425358 (rank : 29) | θ value | 1.38499e-08 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.420568 (rank : 30) | θ value | 4.92598e-06 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.419672 (rank : 31) | θ value | 4.92598e-06 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.419352 (rank : 32) | θ value | 6.43352e-06 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.392472 (rank : 33) | θ value | 2.44474e-05 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.360067 (rank : 34) | θ value | 0.000461057 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.357293 (rank : 35) | θ value | 9.29e-05 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.307923 (rank : 36) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.304579 (rank : 37) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.301695 (rank : 38) | θ value | 0.0193708 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
BRWD1_HUMAN
|
||||||
| NC score | 0.287447 (rank : 39) | θ value | 3.41135e-07 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.287085 (rank : 40) | θ value | 0.00298849 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.280659 (rank : 41) | θ value | 0.0193708 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.278041 (rank : 42) | θ value | 3.19293e-05 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.268944 (rank : 43) | θ value | 1.29631e-06 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.262291 (rank : 44) | θ value | 1.43324e-05 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.214540 (rank : 45) | θ value | 2.88788e-06 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.213950 (rank : 46) | θ value | 4.92598e-06 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.204374 (rank : 47) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.202425 (rank : 48) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.125549 (rank : 49) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.113053 (rank : 50) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.111601 (rank : 51) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.111138 (rank : 52) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.107397 (rank : 53) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.106656 (rank : 54) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.106639 (rank : 55) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.106231 (rank : 56) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.105816 (rank : 57) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.103421 (rank : 58) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.103302 (rank : 59) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.102554 (rank : 60) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.102125 (rank : 61) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.101865 (rank : 62) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.101616 (rank : 63) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.101207 (rank : 64) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.099703 (rank : 65) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.096360 (rank : 66) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.094187 (rank : 67) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.086858 (rank : 68) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.086221 (rank : 69) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.081889 (rank : 70) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.080537 (rank : 71) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.077944 (rank : 72) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.075264 (rank : 73) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.074953 (rank : 74) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.074402 (rank : 75) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.073063 (rank : 76) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.071554 (rank : 77) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.071328 (rank : 78) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.070538 (rank : 79) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.069670 (rank : 80) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.067961 (rank : 81) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.067304 (rank : 82) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.067108 (rank : 83) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.067032 (rank : 84) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.066067 (rank : 85) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.065352 (rank : 86) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TTF1_MOUSE
|
||||||
| NC score | 0.065021 (rank : 87) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.064163 (rank : 88) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.062507 (rank : 89) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.062483 (rank : 90) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.062409 (rank : 91) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.060947 (rank : 92) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.058786 (rank : 93) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.058512 (rank : 94) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.058060 (rank : 95) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.057884 (rank : 96) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.057617 (rank : 97) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.056993 (rank : 98) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.055680 (rank : 99) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.055498 (rank : 100) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.055338 (rank : 101) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
WDR63_HUMAN
|
||||||
| NC score | 0.055037 (rank : 102) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWG1, Q96L72, Q96NU4 | Gene names | WDR63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 63 (Testis development protein NYD-SP29). | |||||
|
JHD3A_MOUSE
|
||||||
| NC score | 0.054966 (rank : 103) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.054942 (rank : 104) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.054421 (rank : 105) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
JHD3A_HUMAN
|
||||||
| NC score | 0.054256 (rank : 106) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3C_HUMAN
|
||||||
| NC score | 0.053174 (rank : 107) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.052413 (rank : 108) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
MTF2_HUMAN
|
||||||
| NC score | 0.051607 (rank : 109) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_MOUSE
|
||||||
| NC score | 0.051197 (rank : 110) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.050488 (rank : 111) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
CF188_MOUSE
|
||||||
| NC score | 0.048294 (rank : 112) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R100, Q8C3G8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf188 homolog. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.045856 (rank : 113) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.041130 (rank : 114) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.032378 (rank : 115) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.030151 (rank : 116) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
CM003_MOUSE
|
||||||
| NC score | 0.030114 (rank : 117) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C263 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3 homolog. | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.025664 (rank : 118) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
K0408_HUMAN
|
||||||
| NC score | 0.025402 (rank : 119) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZU52, O43158, Q5TF20, Q7L2M2 | Gene names | KIAA0408 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0408. | |||||
|
MDM2_MOUSE
|
||||||
| NC score | 0.023693 (rank : 120) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23804, Q61040, Q64330 | Gene names | Mdm2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein). | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.023650 (rank : 121) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
CF188_HUMAN
|
||||||
| NC score | 0.021891 (rank : 122) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5C1 | Gene names | C6orf188 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf188. | |||||
|
PCDG2_HUMAN
|
||||||
| NC score | 0.020512 (rank : 123) | θ value | 0.0431538 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5H1, Q9Y5D5 | Gene names | PCDHGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A2 precursor (PCDH-gamma-A2). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.019550 (rank : 124) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.019496 (rank : 125) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
ITF2_HUMAN
|
||||||
| NC score | 0.018993 (rank : 126) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.018959 (rank : 127) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.018593 (rank : 128) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.016497 (rank : 129) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.015968 (rank : 130) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
PCDGC_HUMAN
|
||||||
| NC score | 0.015211 (rank : 131) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
RXRA_MOUSE
|
||||||
| NC score | 0.014900 (rank : 132) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28700 | Gene names | Rxra, Nr2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
PCDG1_HUMAN
|
||||||
| NC score | 0.014835 (rank : 133) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5H4, Q9Y5D6 | Gene names | PCDHGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A1 precursor (PCDH-gamma-A1). | |||||
|
RXRA_HUMAN
|
||||||
| NC score | 0.014208 (rank : 134) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
PCDG7_HUMAN
|
||||||
| NC score | 0.014138 (rank : 135) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
PCDG3_HUMAN
|
||||||
| NC score | 0.013802 (rank : 136) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5H0, Q9Y5D4 | Gene names | PCDHGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A3 precursor (PCDH-gamma-A3). | |||||
|
PCDGJ_HUMAN
|
||||||
| NC score | 0.013461 (rank : 137) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5F8, Q9UN63 | Gene names | PCDHGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B7 precursor (PCDH-gamma-B7). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.012695 (rank : 138) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.010999 (rank : 139) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
DUS3L_HUMAN
|
||||||
| NC score | 0.010987 (rank : 140) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.010880 (rank : 141) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
HBS1L_HUMAN
|
||||||
| NC score | 0.009435 (rank : 142) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.009374 (rank : 143) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.009335 (rank : 144) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
SEPT4_MOUSE
|
||||||
| NC score | 0.009301 (rank : 145) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28661, Q5ND15 | Gene names | Sept4, Pnutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-4 (Peanut-like protein 2) (Brain protein H5). | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.009023 (rank : 146) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
TBCD1_HUMAN
|
||||||
| NC score | 0.008477 (rank : 147) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.008092 (rank : 148) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CC14B_HUMAN
|
||||||
| NC score | 0.006333 (rank : 149) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.005618 (rank : 150) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.005299 (rank : 151) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
LMNB1_HUMAN
|
||||||
| NC score | 0.002604 (rank : 152) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
PKN3_MOUSE
|
||||||
| NC score | -0.003825 (rank : 153) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||