Please be patient as the page loads
|
TAF1_HUMAN
|
||||||
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TAF1L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985583 (rank : 2) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 3) | NC score | 0.566963 (rank : 6) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 4) | NC score | 0.584216 (rank : 4) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
PB1_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 5) | NC score | 0.547220 (rank : 12) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 6) | NC score | 0.581118 (rank : 5) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 7) | NC score | 0.449113 (rank : 26) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PCAF_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 8) | NC score | 0.597178 (rank : 3) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 9) | NC score | 0.467406 (rank : 21) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 10) | NC score | 0.563868 (rank : 8) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 11) | NC score | 0.559604 (rank : 9) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 12) | NC score | 0.555661 (rank : 10) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 13) | NC score | 0.474974 (rank : 20) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 14) | NC score | 0.415551 (rank : 31) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 15) | NC score | 0.399627 (rank : 34) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 16) | NC score | 0.454596 (rank : 24) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 17) | NC score | 0.503623 (rank : 17) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 18) | NC score | 0.500925 (rank : 18) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BRDT_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 19) | NC score | 0.547300 (rank : 11) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 20) | NC score | 0.427829 (rank : 28) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 21) | NC score | 0.566557 (rank : 7) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 22) | NC score | 0.461027 (rank : 23) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BRD7_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 23) | NC score | 0.527994 (rank : 14) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD7_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 24) | NC score | 0.535102 (rank : 13) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRWD1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 25) | NC score | 0.288989 (rank : 41) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 26) | NC score | 0.425578 (rank : 30) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 27) | NC score | 0.444849 (rank : 27) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 28) | NC score | 0.259459 (rank : 45) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 29) | NC score | 0.452369 (rank : 25) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 30) | NC score | 0.425684 (rank : 29) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 31) | NC score | 0.404111 (rank : 33) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 32) | NC score | 0.408618 (rank : 32) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
BRD9_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 33) | NC score | 0.525218 (rank : 15) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
ATAD2_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 34) | NC score | 0.257247 (rank : 46) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 35) | NC score | 0.374093 (rank : 37) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 36) | NC score | 0.497396 (rank : 19) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 37) | NC score | 0.355745 (rank : 38) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BRD9_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 38) | NC score | 0.523453 (rank : 16) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 39) | NC score | 0.278703 (rank : 42) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 40) | NC score | 0.462448 (rank : 22) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 41) | NC score | 0.379542 (rank : 36) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 42) | NC score | 0.381435 (rank : 35) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 43) | NC score | 0.340487 (rank : 39) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 44) | NC score | 0.335193 (rank : 40) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 45) | NC score | 0.183175 (rank : 49) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 46) | NC score | 0.183527 (rank : 48) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 47) | NC score | 0.278356 (rank : 44) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 48) | NC score | 0.278396 (rank : 43) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 49) | NC score | 0.059557 (rank : 97) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 50) | NC score | 0.050743 (rank : 113) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
LY10_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 51) | NC score | 0.202731 (rank : 47) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 52) | NC score | 0.047900 (rank : 116) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 53) | NC score | 0.047001 (rank : 117) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 0.21417 (rank : 54) | NC score | 0.052469 (rank : 111) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 0.21417 (rank : 55) | NC score | 0.035434 (rank : 137) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 56) | NC score | 0.035853 (rank : 136) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CEP72_MOUSE
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.054886 (rank : 107) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D3R3, Q8BM53, Q9D0B7 | Gene names | Cep72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.025402 (rank : 153) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.022311 (rank : 159) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.027050 (rank : 150) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
SUHW4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.007522 (rank : 180) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
WRN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.023699 (rank : 156) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.051039 (rank : 112) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.029394 (rank : 147) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.042603 (rank : 121) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.055469 (rank : 105) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PPCT_MOUSE
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.045565 (rank : 119) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53808, Q9QZX0, Q9R058 | Gene names | Pctp, Stard2 | |||
|
Domain Architecture |
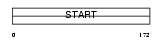
|
|||||
| Description | Phosphatidylcholine transfer protein (PC-TP) (StAR-related lipid transfer protein 2) (StARD2) (START domain-containing protein 2). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.042594 (rank : 122) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.039997 (rank : 126) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.049092 (rank : 115) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RASK_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.005835 (rank : 186) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32883, P04200, P08643 | Gene names | Kras, Kras2 | |||
|
Domain Architecture |
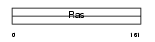
|
|||||
| Description | GTPase KRas (K-Ras 2) (Ki-Ras) (c-K-ras) (c-Ki-ras). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.049174 (rank : 114) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
WDHD1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.027769 (rank : 148) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
UBP13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.026265 (rank : 151) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.032233 (rank : 140) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GAS8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.034529 (rank : 138) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
RASK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.005154 (rank : 188) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01116, P01118, Q96D10 | Gene names | KRAS, KRAS2, RASK2 | |||
|
Domain Architecture |
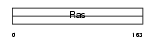
|
|||||
| Description | GTPase KRas (K-Ras 2) (Ki-Ras) (c-K-ras) (c-Ki-ras). | |||||
|
TEX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.037026 (rank : 134) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.027264 (rank : 149) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.042543 (rank : 123) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
JIP3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.031218 (rank : 143) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
K0831_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.038143 (rank : 131) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.038084 (rank : 132) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.098756 (rank : 60) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.092631 (rank : 61) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PVRL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.021735 (rank : 160) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32507, Q62096 | Gene names | Pvrl2, Mph, Pvr, Pvs | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 2 precursor (Murine herpesvirus entry protein B) (mHveB) (Nectin-2) (Poliovirus receptor homolog) (CD112 antigen). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.017651 (rank : 166) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.031281 (rank : 142) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.007003 (rank : 181) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.056897 (rank : 103) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.031507 (rank : 141) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
STX1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.019235 (rank : 163) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15531 | Gene names | STX1B1, STX1B | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1B1 (Syntaxin 1B). | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.042466 (rank : 124) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.039536 (rank : 128) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GRIP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.009354 (rank : 178) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
HS70L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.005336 (rank : 187) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.003019 (rank : 189) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
TNIP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.038217 (rank : 130) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.069607 (rank : 90) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.037373 (rank : 133) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.040320 (rank : 125) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MYO1F_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.009003 (rank : 179) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00160, Q8WWN7 | Gene names | MYO1F | |||
|
Domain Architecture |

|
|||||
| Description | Myosin If (Myosin-IE). | |||||
|
NOL6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.024074 (rank : 155) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H6R4, Q5T5M3, Q5T5M4, Q7L4G6, Q8N6I0, Q8TEY9, Q8TEZ0, Q8TEZ1, Q9H675 | Gene names | NOL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.018908 (rank : 164) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.015114 (rank : 169) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
SDS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.036989 (rank : 135) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BR65, Q3UC92, Q6P6K1, Q7TNT0, Q8BRR7, Q8K5B4 | Gene names | Suds3, Sds3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sin3 histone deacetylase corepressor complex component SDS3 (Suppressor of defective silencing 3 protein homolog). | |||||
|
TBX3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.006018 (rank : 184) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.012828 (rank : 173) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.046497 (rank : 118) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.030787 (rank : 144) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
ERCC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.023216 (rank : 157) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92889, O00140, Q8TD83 | Gene names | ERCC4, ERCC11, XPF | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4) (DNA-repair protein complementing XP-F cells) (Xeroderma pigmentosum group F-complementing protein). | |||||
|
ERCC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.020735 (rank : 162) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
HOME3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.022975 (rank : 158) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NSC5, O14580, O95350, Q9NSB9, Q9NSC0 | Gene names | HOMER3 | |||
|
Domain Architecture |
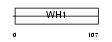
|
|||||
| Description | Homer protein homolog 3 (Homer-3). | |||||
|
K2C8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.012214 (rank : 174) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.021000 (rank : 161) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
MPP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.005869 (rank : 185) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13368 | Gene names | MPP3, DLG3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Discs large homolog 3). | |||||
|
P85B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.015117 (rank : 168) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
P85B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.015579 (rank : 167) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
REST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.039025 (rank : 129) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RRFM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.030497 (rank : 145) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D6S7 | Gene names | Mrrf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosome recycling factor, mitochondrial precursor. | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.024094 (rank : 154) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
CLH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.011267 (rank : 176) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
DNMT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.018006 (rank : 165) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14717, O43669 | Gene names | DNMT2 | |||
|
Domain Architecture |
|
|||||
| Description | tRNA (cytosine-5-)-methyltransferase (EC 2.1.1.29) (DNA (cytosine-5)- methyltransferase-like protein 2) (Dnmt2) (DNA methyltransferase homolog HsaIIP) (DNA MTase homolog HsaIIP) (M.HsaIIP) (PuMet). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.006745 (rank : 182) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.039589 (rank : 127) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.033400 (rank : 139) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
MOR2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.013545 (rank : 172) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
NC6IP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.014124 (rank : 170) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RS0, Q5GH23, Q8TDG9, Q96QU3, Q9H5V3 | Gene names | NCOA6IP, PIMT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT) (CLL-associated antigen KW-2) (Hepatocellular carcinoma-associated antigen 137) (HCA137). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.029923 (rank : 146) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
RRFM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.026086 (rank : 152) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96E11, Q5RKT1, Q5T7T0, Q5T7T1, Q5T7T2, Q5T7T3, Q5T7T4, Q5T7T5 | Gene names | MRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosome recycling factor, mitochondrial precursor. | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.006222 (rank : 183) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.010425 (rank : 177) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.043238 (rank : 120) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
ZBT10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | -0.002409 (rank : 190) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DT7, Q86W96, Q8IXI9, Q96MH9 | Gene names | ZBTB10, RINZF, RINZFC | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 10 (Zinc finger protein RIN ZF). | |||||
|
ZFYV1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.013979 (rank : 171) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
ZFYV1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.012065 (rank : 175) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810J8 | Gene names | Zfyve1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1. | |||||
|
AF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.058088 (rank : 102) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.060013 (rank : 96) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.066314 (rank : 93) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.141347 (rank : 50) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.138152 (rank : 51) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054510 (rank : 108) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.078815 (rank : 73) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.077874 (rank : 77) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.077916 (rank : 76) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.078754 (rank : 74) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.072771 (rank : 85) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.077630 (rank : 78) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.066389 (rank : 92) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.055526 (rank : 104) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.055103 (rank : 106) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.062457 (rank : 94) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.058638 (rank : 100) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JADE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.076686 (rank : 80) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.076829 (rank : 79) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.074800 (rank : 82) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.074524 (rank : 83) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.078197 (rank : 75) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.080859 (rank : 72) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.071617 (rank : 87) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.072822 (rank : 84) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.071217 (rank : 88) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.084626 (rank : 68) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.081266 (rank : 71) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.072467 (rank : 86) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.075409 (rank : 81) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.126300 (rank : 56) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.132666 (rank : 54) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.100355 (rank : 59) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.091938 (rank : 62) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.087804 (rank : 64) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.086519 (rank : 65) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.106977 (rank : 57) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.106779 (rank : 58) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.085400 (rank : 67) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.083836 (rank : 70) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.052952 (rank : 109) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.052503 (rank : 110) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.086338 (rank : 66) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.084130 (rank : 69) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.130254 (rank : 55) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SP110_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.089743 (rank : 63) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.135028 (rank : 52) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.133167 (rank : 53) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
TRI45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.060218 (rank : 95) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H8W5, Q5T2K4, Q5T2K5, Q8IYV6 | Gene names | TRIM45, RNF99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45 (RING finger protein 99). | |||||
|
TRI45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.059433 (rank : 98) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PFY8, Q8BVT5 | Gene names | Trim45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45. | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.066994 (rank : 91) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.069996 (rank : 89) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.058558 (rank : 101) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.058983 (rank : 99) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
TAF1L_HUMAN
|
||||||
| NC score | 0.985583 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
PCAF_HUMAN
|
||||||
| NC score | 0.597178 (rank : 3) | θ value | 6.64225e-11 (rank : 8) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.584216 (rank : 4) | θ value | 4.16044e-13 (rank : 4) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.581118 (rank : 5) | θ value | 7.09661e-13 (rank : 6) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.566963 (rank : 6) | θ value | 1.68911e-14 (rank : 3) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.566557 (rank : 7) | θ value | 8.11959e-09 (rank : 21) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.563868 (rank : 8) | θ value | 1.133e-10 (rank : 10) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.559604 (rank : 9) | θ value | 1.133e-10 (rank : 11) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.555661 (rank : 10) | θ value | 2.52405e-10 (rank : 12) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
BRDT_MOUSE
|
||||||
| NC score | 0.547300 (rank : 11) | θ value | 6.21693e-09 (rank : 19) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.547220 (rank : 12) | θ value | 4.16044e-13 (rank : 5) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.535102 (rank : 13) | θ value | 2.36244e-08 (rank : 24) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| NC score | 0.527994 (rank : 14) | θ value | 1.80886e-08 (rank : 23) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD9_HUMAN
|
||||||
| NC score | 0.525218 (rank : 15) | θ value | 5.81887e-07 (rank : 33) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD9_MOUSE
|
||||||
| NC score | 0.523453 (rank : 16) | θ value | 9.92553e-07 (rank : 38) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.503623 (rank : 17) | θ value | 4.76016e-09 (rank : 17) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.500925 (rank : 18) | θ value | 4.76016e-09 (rank : 18) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.497396 (rank : 19) | θ value | 7.59969e-07 (rank : 36) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.474974 (rank : 20) | θ value | 2.13673e-09 (rank : 13) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.467406 (rank : 21) | θ value | 8.67504e-11 (rank : 9) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.462448 (rank : 22) | θ value | 6.43352e-06 (rank : 40) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.461027 (rank : 23) | θ value | 1.38499e-08 (rank : 22) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.454596 (rank : 24) | θ value | 3.64472e-09 (rank : 16) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.452369 (rank : 25) | θ value | 5.26297e-08 (rank : 29) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.449113 (rank : 26) | θ value | 2.28291e-11 (rank : 7) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.444849 (rank : 27) | θ value | 4.0297e-08 (rank : 27) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.427829 (rank : 28) | θ value | 6.21693e-09 (rank : 20) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.425684 (rank : 29) | θ value | 5.26297e-08 (rank : 30) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.425578 (rank : 30) | θ value | 4.0297e-08 (rank : 26) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.415551 (rank : 31) | θ value | 2.13673e-09 (rank : 14) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.408618 (rank : 32) | θ value | 3.41135e-07 (rank : 32) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.404111 (rank : 33) | θ value | 1.99992e-07 (rank : 31) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.399627 (rank : 34) | θ value | 2.79066e-09 (rank : 15) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.381435 (rank : 35) | θ value | 1.09739e-05 (rank : 42) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.379542 (rank : 36) | θ value | 8.40245e-06 (rank : 41) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.374093 (rank : 37) | θ value | 7.59969e-07 (rank : 35) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.355745 (rank : 38) | θ value | 7.59969e-07 (rank : 37) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.340487 (rank : 39) | θ value | 3.19293e-05 (rank : 43) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.335193 (rank : 40) | θ value | 4.1701e-05 (rank : 44) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
BRWD1_HUMAN
|
||||||
| NC score | 0.288989 (rank : 41) | θ value | 2.36244e-08 (rank : 25) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NSI6, O43721, Q5R2V0, Q5R2V1, Q8TCV3, Q96QG9, Q96QH0, Q9NUK1 | Gene names | BRWD1, C21orf107, WDR9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.278703 (rank : 42) | θ value | 9.92553e-07 (rank : 39) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.278396 (rank : 43) | θ value | 0.00298849 (rank : 48) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.278356 (rank : 44) | θ value | 0.00298849 (rank : 47) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.259459 (rank : 45) | θ value | 5.26297e-08 (rank : 28) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
ATAD2_MOUSE
|
||||||
| NC score | 0.257247 (rank : 46) | θ value | 7.59969e-07 (rank : 34) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8CDM1 | Gene names | Atad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.202731 (rank : 47) | θ value | 0.0961366 (rank : 51) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.183527 (rank : 48) | θ value | 0.00102713 (rank : 46) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.183175 (rank : 49) | θ value | 0.000786445 (rank : 45) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.141347 (rank : 50) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.138152 (rank : 51) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.135028 (rank : 52) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.133167 (rank : 53) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.132666 (rank : 54) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.130254 (rank : 55) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.126300 (rank : 56) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.106977 (rank : 57) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.106779 (rank : 58) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.100355 (rank : 59) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.098756 (rank : 60) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.092631 (rank : 61) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.091938 (rank : 62) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.089743 (rank : 63) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.087804 (rank : 64) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.086519 (rank : 65) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.086338 (rank : 66) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.085400 (rank : 67) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.084626 (rank : 68) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.084130 (rank : 69) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.083836 (rank : 70) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.081266 (rank : 71) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.080859 (rank : 72) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.078815 (rank : 73) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.078754 (rank : 74) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.078197 (rank : 75) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.077916 (rank : 76) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.077874 (rank : 77) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.077630 (rank : 78) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.076829 (rank : 79) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.076686 (rank : 80) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.075409 (rank : 81) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.074800 (rank : 82) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.074524 (rank : 83) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.072822 (rank : 84) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.072771 (rank : 85) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.072467 (rank : 86) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.071617 (rank : 87) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.071217 (rank : 88) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.069996 (rank : 89) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.069607 (rank : 90) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.066994 (rank : 91) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.066389 (rank : 92) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.066314 (rank : 93) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.062457 (rank : 94) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
TRI45_HUMAN
|
||||||
| NC score | 0.060218 (rank : 95) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H8W5, Q5T2K4, Q5T2K5, Q8IYV6 | Gene names | TRIM45, RNF99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45 (RING finger protein 99). | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.060013 (rank : 96) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.059557 (rank : 97) | θ value | 0.0330416 (rank : 49) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TRI45_MOUSE
|
||||||
| NC score | 0.059433 (rank : 98) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PFY8, Q8BVT5 | Gene names | Trim45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45. | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.058983 (rank : 99) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
JAD1D_MOUSE
|
||||||
| NC score | 0.058638 (rank : 100) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.058558 (rank : 101) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.058088 (rank : 102) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.056897 (rank : 103) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.055526 (rank : 104) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.055469 (rank : 105) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.055103 (rank : 106) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
CEP72_MOUSE
|
||||||
| NC score | 0.054886 (rank : 107) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D3R3, Q8BM53, Q9D0B7 | Gene names | Cep72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.054510 (rank : 108) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.052952 (rank : 109) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.052503 (rank : 110) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.052469 (rank : 111) | θ value | 0.21417 (rank : 54) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.051039 (rank : 112) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.050743 (rank : 113) | θ value | 0.0961366 (rank : 50) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.049174 (rank : 114) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.049092 (rank : 115) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.047900 (rank : 116) | θ value | 0.0961366 (rank : 52) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.047001 (rank : 117) | θ value | 0.125558 (rank : 53) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.046497 (rank : 118) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
PPCT_MOUSE
|
||||||
| NC score | 0.045565 (rank : 119) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53808, Q9QZX0, Q9R058 | Gene names | Pctp, Stard2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylcholine transfer protein (PC-TP) (StAR-related lipid transfer protein 2) (StARD2) (START domain-containing protein 2). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.043238 (rank : 120) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.042603 (rank : 121) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.042594 (rank : 122) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.042543 (rank : 123) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.042466 (rank : 124) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.040320 (rank : 125) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.039997 (rank : 126) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.039589 (rank : 127) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.039536 (rank : 128) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.039025 (rank : 129) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
TNIP3_HUMAN
|
||||||
| NC score | 0.038217 (rank : 130) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
K0831_MOUSE
|
||||||
| NC score | 0.038143 (rank : 131) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.038084 (rank : 132) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.037373 (rank : 133) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
TEX2_HUMAN
|
||||||
| NC score | 0.037026 (rank : 134) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
SDS3_MOUSE
|
||||||
| NC score | 0.036989 (rank : 135) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BR65, Q3UC92, Q6P6K1, Q7TNT0, Q8BRR7, Q8K5B4 | Gene names | Suds3, Sds3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sin3 histone deacetylase corepressor complex component SDS3 (Suppressor of defective silencing 3 protein homolog). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.035853 (rank : 136) | θ value | 0.279714 (rank : 56) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.035434 (rank : 137) | θ value | 0.21417 (rank : 55) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.034529 (rank : 138) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.033400 (rank : 139) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.032233 (rank : 140) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.031507 (rank : 141) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.031281 (rank : 142) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
JIP3_MOUSE
|
||||||
| NC score | 0.031218 (rank : 143) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ESN9, Q99KU7, Q9EQD8, Q9ESN7, Q9ESN8, Q9ESP0, Q9JLH2, Q9JLH3, Q9R0U7 | Gene names | Mapk8ip3, Jip3, Jsap1, Syd2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3) (JNK/SAPK-associated protein 1) (JSAP1) (Sunday driver 2). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.030787 (rank : 144) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
RRFM_MOUSE
|
||||||
| NC score | 0.030497 (rank : 145) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D6S7 | Gene names | Mrrf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosome recycling factor, mitochondrial precursor. | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.029923 (rank : 146) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.029394 (rank : 147) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
WDHD1_HUMAN
|
||||||
| NC score | 0.027769 (rank : 148) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.027264 (rank : 149) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.027050 (rank : 150) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
UBP13_HUMAN
|
||||||
| NC score | 0.026265 (rank : 151) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92995 | Gene names | USP13, ISOT3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 13 (EC 3.1.2.15) (Ubiquitin thioesterase 13) (Ubiquitin-specific-processing protease 13) (Deubiquitinating enzyme 13) (Isopeptidase T-3) (ISOT-3). | |||||
|
RRFM_HUMAN
|
||||||
| NC score | 0.026086 (rank : 152) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96E11, Q5RKT1, Q5T7T0, Q5T7T1, Q5T7T2, Q5T7T3, Q5T7T4, Q5T7T5 | Gene names | MRRF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosome recycling factor, mitochondrial precursor. | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.025402 (rank : 153) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.024094 (rank : 154) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
NOL6_HUMAN
|
||||||
| NC score | 0.024074 (rank : 155) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H6R4, Q5T5M3, Q5T5M4, Q7L4G6, Q8N6I0, Q8TEY9, Q8TEZ0, Q8TEZ1, Q9H675 | Gene names | NOL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 6 (Nucleolar RNA-associated protein) (Nrap). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.023699 (rank : 156) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
ERCC4_HUMAN
|
||||||
| NC score | 0.023216 (rank : 157) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92889, O00140, Q8TD83 | Gene names | ERCC4, ERCC11, XPF | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4) (DNA-repair protein complementing XP-F cells) (Xeroderma pigmentosum group F-complementing protein). | |||||
|
HOME3_HUMAN
|
||||||
| NC score | 0.022975 (rank : 158) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NSC5, O14580, O95350, Q9NSB9, Q9NSC0 | Gene names | HOMER3 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 3 (Homer-3). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.022311 (rank : 159) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PVRL2_MOUSE
|
||||||
| NC score | 0.021735 (rank : 160) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32507, Q62096 | Gene names | Pvrl2, Mph, Pvr, Pvs | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 2 precursor (Murine herpesvirus entry protein B) (mHveB) (Nectin-2) (Poliovirus receptor homolog) (CD112 antigen). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.021000 (rank : 161) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
ERCC4_MOUSE
|
||||||
| NC score | 0.020735 (rank : 162) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZD4, O54810, Q8R0I3 | Gene names | Ercc4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4). | |||||
|
STX1B_HUMAN
|
||||||
| NC score | 0.019235 (rank : 163) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15531 | Gene names | STX1B1, STX1B | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-1B1 (Syntaxin 1B). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.018908 (rank : 164) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
DNMT2_HUMAN
|
||||||
| NC score | 0.018006 (rank : 165) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14717, O43669 | Gene names | DNMT2 | |||
|
Domain Architecture |
|
|||||
| Description | tRNA (cytosine-5-)-methyltransferase (EC 2.1.1.29) (DNA (cytosine-5)- methyltransferase-like protein 2) (Dnmt2) (DNA methyltransferase homolog HsaIIP) (DNA MTase homolog HsaIIP) (M.HsaIIP) (PuMet). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.017651 (rank : 166) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.015579 (rank : 167) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.015117 (rank : 168) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.015114 (rank : 169) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
NC6IP_HUMAN
|
||||||
| NC score | 0.014124 (rank : 170) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RS0, Q5GH23, Q8TDG9, Q96QU3, Q9H5V3 | Gene names | NCOA6IP, PIMT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA methyltransferase NCOA6IP (EC 2.1.1.-) (Nuclear receptor coactivator 6-interacting protein) (PRIP-interacting protein with methyltransferase motif) (PIPMT) (PIMT) (CLL-associated antigen KW-2) (Hepatocellular carcinoma-associated antigen 137) (HCA137). | |||||
|
ZFYV1_HUMAN
|
||||||
| NC score | 0.013979 (rank : 171) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
MOR2A_MOUSE
|
||||||
| NC score | 0.013545 (rank : 172) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.012828 (rank : 173) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
K2C8_HUMAN
|
||||||
| NC score | 0.012214 (rank : 174) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
ZFYV1_MOUSE
|
||||||
| NC score | 0.012065 (rank : 175) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810J8 | Gene names | Zfyve1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1. | |||||
|
CLH2_HUMAN
|
||||||
| NC score | 0.011267 (rank : 176) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.010425 (rank : 177) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
GRIP1_MOUSE
|
||||||
| NC score | 0.009354 (rank : 178) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
MYO1F_HUMAN
|
||||||
| NC score | 0.009003 (rank : 179) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00160, Q8WWN7 | Gene names | MYO1F | |||
|
Domain Architecture |

|
|||||
| Description | Myosin If (Myosin-IE). | |||||
|
SUHW4_MOUSE
|
||||||
| NC score | 0.007522 (rank : 180) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.007003 (rank : 181) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.006745 (rank : 182) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.006222 (rank : 183) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
TBX3_HUMAN
|
||||||
| NC score | 0.006018 (rank : 184) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
MPP3_HUMAN
|
||||||
| NC score | 0.005869 (rank : 185) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13368 | Gene names | MPP3, DLG3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Discs large homolog 3). | |||||
|
RASK_MOUSE
|
||||||
| NC score | 0.005835 (rank : 186) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32883, P04200, P08643 | Gene names | Kras, Kras2 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase KRas (K-Ras 2) (Ki-Ras) (c-K-ras) (c-Ki-ras). | |||||
|
HS70L_HUMAN
|
||||||
| NC score | 0.005336 (rank : 187) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P34931, O75634, Q2HXR3, Q8NE72, Q96QC9, Q9UQM1 | Gene names | HSPA1L | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 1L (Heat shock 70 kDa protein 1-like) (Heat shock 70 kDa protein 1-Hom) (HSP70-Hom). | |||||
|
RASK_HUMAN
|
||||||
| NC score | 0.005154 (rank : 188) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01116, P01118, Q96D10 | Gene names | KRAS, KRAS2, RASK2 | |||
|
Domain Architecture |

|
|||||
| Description | GTPase KRas (K-Ras 2) (Ki-Ras) (c-K-ras) (c-Ki-ras). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.003019 (rank : 189) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
ZBT10_HUMAN
|
||||||
| NC score | -0.002409 (rank : 190) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 136 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DT7, Q86W96, Q8IXI9, Q96MH9 | Gene names | ZBTB10, RINZF, RINZFC | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 10 (Zinc finger protein RIN ZF). | |||||