Please be patient as the page loads
|
WRN_HUMAN
|
||||||
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
WRN_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.960445 (rank : 2) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | 9.43692e-66 (rank : 3) | NC score | 0.873212 (rank : 4) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 2.10232e-65 (rank : 4) | NC score | 0.876514 (rank : 3) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
BLM_MOUSE
|
||||||
| θ value | 3.84025e-59 (rank : 5) | NC score | 0.853674 (rank : 5) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| θ value | 1.61343e-57 (rank : 6) | NC score | 0.846533 (rank : 6) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 1.51013e-55 (rank : 7) | NC score | 0.820234 (rank : 7) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 3.61106e-41 (rank : 8) | NC score | 0.808735 (rank : 8) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 5.21438e-40 (rank : 9) | NC score | 0.799419 (rank : 9) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 10) | NC score | 0.518026 (rank : 13) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 11) | NC score | 0.517809 (rank : 14) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 12) | NC score | 0.498683 (rank : 27) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 13) | NC score | 0.503051 (rank : 22) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
EXDL2_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 14) | NC score | 0.254095 (rank : 78) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVH0, Q8N3D3 | Gene names | EXDL2, C14orf114 | |||
|
Domain Architecture |
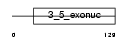
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
EXDL2_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 15) | NC score | 0.250269 (rank : 79) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEG4 | Gene names | Exdl2 | |||
|
Domain Architecture |
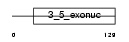
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 16) | NC score | 0.501528 (rank : 24) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 17) | NC score | 0.522558 (rank : 11) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 18) | NC score | 0.522630 (rank : 10) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 19) | NC score | 0.499734 (rank : 26) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 20) | NC score | 0.500361 (rank : 25) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 21) | NC score | 0.504129 (rank : 17) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 22) | NC score | 0.494201 (rank : 30) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 23) | NC score | 0.493998 (rank : 31) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 24) | NC score | 0.517767 (rank : 15) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 25) | NC score | 0.518247 (rank : 12) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 26) | NC score | 0.506150 (rank : 16) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 27) | NC score | 0.503053 (rank : 21) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 28) | NC score | 0.503859 (rank : 19) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 29) | NC score | 0.503758 (rank : 20) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 30) | NC score | 0.474014 (rank : 43) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 31) | NC score | 0.479053 (rank : 38) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 32) | NC score | 0.479053 (rank : 39) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 33) | NC score | 0.491591 (rank : 32) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 34) | NC score | 0.470921 (rank : 47) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 35) | NC score | 0.468722 (rank : 50) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 36) | NC score | 0.479660 (rank : 36) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 37) | NC score | 0.479660 (rank : 37) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 38) | NC score | 0.503986 (rank : 18) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 39) | NC score | 0.502209 (rank : 23) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 40) | NC score | 0.498627 (rank : 28) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 41) | NC score | 0.498458 (rank : 29) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 42) | NC score | 0.470188 (rank : 49) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 43) | NC score | 0.484959 (rank : 35) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 44) | NC score | 0.488640 (rank : 33) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 45) | NC score | 0.488400 (rank : 34) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 46) | NC score | 0.477661 (rank : 41) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 47) | NC score | 0.474744 (rank : 42) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 48) | NC score | 0.470393 (rank : 48) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 49) | NC score | 0.478780 (rank : 40) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 50) | NC score | 0.468102 (rank : 52) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 51) | NC score | 0.472446 (rank : 45) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 52) | NC score | 0.472215 (rank : 46) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 53) | NC score | 0.430809 (rank : 66) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 54) | NC score | 0.449408 (rank : 58) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 55) | NC score | 0.446955 (rank : 59) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 56) | NC score | 0.449890 (rank : 57) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 57) | NC score | 0.457015 (rank : 56) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 58) | NC score | 0.178648 (rank : 85) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 59) | NC score | 0.460563 (rank : 54) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 60) | NC score | 0.460418 (rank : 55) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 61) | NC score | 0.423939 (rank : 72) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 62) | NC score | 0.430260 (rank : 67) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 63) | NC score | 0.429198 (rank : 69) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 64) | NC score | 0.430021 (rank : 68) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
U520_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 65) | NC score | 0.198409 (rank : 84) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 66) | NC score | 0.446347 (rank : 60) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 67) | NC score | 0.426105 (rank : 70) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 68) | NC score | 0.421894 (rank : 73) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
MTO1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 69) | NC score | 0.044484 (rank : 99) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923Z3, Q9D2Q5 | Gene names | Mto1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MTO1 homolog, mitochondrial precursor. | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 70) | NC score | 0.438139 (rank : 63) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 71) | NC score | 0.440995 (rank : 62) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 72) | NC score | 0.468543 (rank : 51) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 73) | NC score | 0.467947 (rank : 53) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 74) | NC score | 0.025891 (rank : 105) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SKIV2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 75) | NC score | 0.121814 (rank : 90) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 0.21417 (rank : 76) | NC score | 0.441229 (rank : 61) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.279714 (rank : 77) | NC score | 0.013175 (rank : 129) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 0.279714 (rank : 78) | NC score | 0.431911 (rank : 65) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 79) | NC score | 0.026433 (rank : 104) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 0.365318 (rank : 80) | NC score | 0.434098 (rank : 64) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 0.365318 (rank : 81) | NC score | 0.014992 (rank : 123) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 82) | NC score | 0.013282 (rank : 127) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 83) | NC score | 0.018825 (rank : 112) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
SK2L2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 84) | NC score | 0.106149 (rank : 91) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P42285, Q8TAG2 | Gene names | SKIV2L2, KIAA0052 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
SK2L2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 85) | NC score | 0.104504 (rank : 92) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZU3 | Gene names | Skiv2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 86) | NC score | 0.023699 (rank : 106) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 87) | NC score | 0.018277 (rank : 114) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.019270 (rank : 111) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.027775 (rank : 102) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
ZW10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.048378 (rank : 98) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54692, Q3TIA5, Q3ULW1, Q921H3 | Gene names | Zw10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 1.06291 (rank : 91) | NC score | 0.015252 (rank : 122) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.036542 (rank : 100) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.015912 (rank : 119) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.415248 (rank : 74) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.208797 (rank : 80) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
IPP4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.029288 (rank : 101) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
ANR52_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.009612 (rank : 135) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.009576 (rank : 136) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.017298 (rank : 116) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.015684 (rank : 120) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.018379 (rank : 113) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.013142 (rank : 130) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.172870 (rank : 86) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.027129 (rank : 103) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
POT15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.013224 (rank : 128) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
TUR8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.055963 (rank : 96) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19473 | Gene names | Trap1a, P1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor rejection antigen P815A. | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.017445 (rank : 115) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
CR034_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.019451 (rank : 110) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.424315 (rank : 71) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.411766 (rank : 75) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.017173 (rank : 117) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.017166 (rank : 118) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.012524 (rank : 131) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.007925 (rank : 137) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.019775 (rank : 109) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.013985 (rank : 126) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5A7, O95422, Q8IX22, Q9BXR2 | Gene names | NUB1, NYREN18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (NY-REN-18 antigen). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.011103 (rank : 133) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
AN32A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.014203 (rank : 124) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.014203 (rank : 125) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.020125 (rank : 108) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
GCC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.015587 (rank : 121) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D4H2, Q8CCR3 | Gene names | Gcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
INVO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.020148 (rank : 107) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
NEBU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.007538 (rank : 138) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |
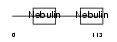
|
|||||
| Description | Nebulin. | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.004061 (rank : 140) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.010186 (rank : 134) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.011330 (rank : 132) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.199851 (rank : 83) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.201951 (rank : 82) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
NBEAL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.003912 (rank : 141) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.002129 (rank : 142) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
RP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.006513 (rank : 139) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |
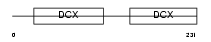
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
ZFP37_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | -0.005283 (rank : 143) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17141, Q62514 | Gene names | Zfp37, Zfp-37 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 37 (Zfp-37) (Male germ cell-specific zinc finger protein). | |||||
|
CC026_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.074635 (rank : 95) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
CC026_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.055196 (rank : 97) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
DDX53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.473056 (rank : 44) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.396331 (rank : 77) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.396880 (rank : 76) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.075358 (rank : 94) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.094545 (rank : 93) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.203050 (rank : 81) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.170861 (rank : 87) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.165871 (rank : 89) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.169434 (rank : 88) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.960445 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.876514 (rank : 3) | θ value | 2.10232e-65 (rank : 4) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.873212 (rank : 4) | θ value | 9.43692e-66 (rank : 3) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.853674 (rank : 5) | θ value | 3.84025e-59 (rank : 5) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.846533 (rank : 6) | θ value | 1.61343e-57 (rank : 6) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.820234 (rank : 7) | θ value | 1.51013e-55 (rank : 7) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.808735 (rank : 8) | θ value | 3.61106e-41 (rank : 8) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.799419 (rank : 9) | θ value | 5.21438e-40 (rank : 9) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.522630 (rank : 10) | θ value | 1.06045e-08 (rank : 18) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.522558 (rank : 11) | θ value | 1.06045e-08 (rank : 17) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.518247 (rank : 12) | θ value | 2.61198e-07 (rank : 25) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.518026 (rank : 13) | θ value | 1.63604e-09 (rank : 10) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.517809 (rank : 14) | θ value | 1.63604e-09 (rank : 11) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.517767 (rank : 15) | θ value | 2.61198e-07 (rank : 24) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.506150 (rank : 16) | θ value | 1.29631e-06 (rank : 26) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.504129 (rank : 17) | θ value | 1.17247e-07 (rank : 21) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.503986 (rank : 18) | θ value | 1.43324e-05 (rank : 38) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.503859 (rank : 19) | θ value | 2.21117e-06 (rank : 28) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.503758 (rank : 20) | θ value | 2.21117e-06 (rank : 29) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.503053 (rank : 21) | θ value | 1.69304e-06 (rank : 27) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.503051 (rank : 22) | θ value | 3.64472e-09 (rank : 13) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.502209 (rank : 23) | θ value | 1.43324e-05 (rank : 39) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.501528 (rank : 24) | θ value | 8.11959e-09 (rank : 16) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.500361 (rank : 25) | θ value | 8.97725e-08 (rank : 20) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.499734 (rank : 26) | θ value | 4.0297e-08 (rank : 19) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.498683 (rank : 27) | θ value | 2.13673e-09 (rank : 12) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.498627 (rank : 28) | θ value | 1.43324e-05 (rank : 40) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.498458 (rank : 29) | θ value | 1.43324e-05 (rank : 41) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.494201 (rank : 30) | θ value | 1.99992e-07 (rank : 22) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.493998 (rank : 31) | θ value | 1.99992e-07 (rank : 23) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.491591 (rank : 32) | θ value | 6.43352e-06 (rank : 33) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.488640 (rank : 33) | θ value | 4.1701e-05 (rank : 44) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.488400 (rank : 34) | θ value | 0.000158464 (rank : 45) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.484959 (rank : 35) | θ value | 4.1701e-05 (rank : 43) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.479660 (rank : 36) | θ value | 1.09739e-05 (rank : 36) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.479660 (rank : 37) | θ value | 1.09739e-05 (rank : 37) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.479053 (rank : 38) | θ value | 4.92598e-06 (rank : 31) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.479053 (rank : 39) | θ value | 4.92598e-06 (rank : 32) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.478780 (rank : 40) | θ value | 0.00102713 (rank : 49) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.477661 (rank : 41) | θ value | 0.000270298 (rank : 46) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.474744 (rank : 42) | θ value | 0.000461057 (rank : 47) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.474014 (rank : 43) | θ value | 2.88788e-06 (rank : 30) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.473056 (rank : 44) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.472446 (rank : 45) | θ value | 0.00102713 (rank : 51) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.472215 (rank : 46) | θ value | 0.00102713 (rank : 52) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.470921 (rank : 47) | θ value | 6.43352e-06 (rank : 34) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.470393 (rank : 48) | θ value | 0.000786445 (rank : 48) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.470188 (rank : 49) | θ value | 2.44474e-05 (rank : 42) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.468722 (rank : 50) | θ value | 8.40245e-06 (rank : 35) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.468543 (rank : 51) | θ value | 0.0961366 (rank : 72) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.468102 (rank : 52) | θ value | 0.00102713 (rank : 50) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.467947 (rank : 53) | θ value | 0.0961366 (rank : 73) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.460563 (rank : 54) | θ value | 0.00869519 (rank : 59) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.460418 (rank : 55) | θ value | 0.00869519 (rank : 60) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.457015 (rank : 56) | θ value | 0.00665767 (rank : 57) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.449890 (rank : 57) | θ value | 0.00665767 (rank : 56) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.449408 (rank : 58) | θ value | 0.00228821 (rank : 54) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.446955 (rank : 59) | θ value | 0.00509761 (rank : 55) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.446347 (rank : 60) | θ value | 0.0193708 (rank : 66) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.441229 (rank : 61) | θ value | 0.21417 (rank : 76) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.440995 (rank : 62) | θ value | 0.0961366 (rank : 71) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.438139 (rank : 63) | θ value | 0.0961366 (rank : 70) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.434098 (rank : 64) | θ value | 0.365318 (rank : 80) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.431911 (rank : 65) | θ value | 0.279714 (rank : 78) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.430809 (rank : 66) | θ value | 0.00175202 (rank : 53) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.430260 (rank : 67) | θ value | 0.0148317 (rank : 62) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.430021 (rank : 68) | θ value | 0.0148317 (rank : 64) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.429198 (rank : 69) | θ value | 0.0148317 (rank : 63) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.426105 (rank : 70) | θ value | 0.0193708 (rank : 67) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.424315 (rank : 71) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.423939 (rank : 72) | θ value | 0.0113563 (rank : 61) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.421894 (rank : 73) | θ value | 0.0736092 (rank : 68) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.415248 (rank : 74) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.411766 (rank : 75) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.396880 (rank : 76) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.396331 (rank : 77) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
EXDL2_HUMAN
|
||||||
| NC score | 0.254095 (rank : 78) | θ value | 3.64472e-09 (rank : 14) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVH0, Q8N3D3 | Gene names | EXDL2, C14orf114 | |||
|
Domain Architecture |
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
EXDL2_MOUSE
|
||||||
| NC score | 0.250269 (rank : 79) | θ value | 3.64472e-09 (rank : 15) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEG4 | Gene names | Exdl2 | |||
|
Domain Architecture |
|
|||||
| Description | Exonuclease 3'-5' domain-like-containing protein 2. | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.208797 (rank : 80) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.203050 (rank : 81) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.201951 (rank : 82) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.199851 (rank : 83) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.198409 (rank : 84) | θ value | 0.0148317 (rank : 65) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.178648 (rank : 85) | θ value | 0.00869519 (rank : 58) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.172870 (rank : 86) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.170861 (rank : 87) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.169434 (rank : 88) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.165871 (rank : 89) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
SKIV2_HUMAN
|
||||||
| NC score | 0.121814 (rank : 90) | θ value | 0.163984 (rank : 75) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15477, O15005, Q12902, Q15476 | Gene names | SKIV2L, DDX13, SKI2W, SKIV2, W | |||
|
Domain Architecture |

|
|||||
| Description | Helicase SKI2W (EC 3.6.1.-) (Helicase-like protein) (HLP). | |||||
|
SK2L2_HUMAN
|
||||||
| NC score | 0.106149 (rank : 91) | θ value | 0.47712 (rank : 84) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P42285, Q8TAG2 | Gene names | SKIV2L2, KIAA0052 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
SK2L2_MOUSE
|
||||||
| NC score | 0.104504 (rank : 92) | θ value | 0.47712 (rank : 85) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZU3 | Gene names | Skiv2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.094545 (rank : 93) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.075358 (rank : 94) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.074635 (rank : 95) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
TUR8_MOUSE
|
||||||
| NC score | 0.055963 (rank : 96) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19473 | Gene names | Trap1a, P1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor rejection antigen P815A. | |||||
|
CC026_MOUSE
|
||||||
| NC score | 0.055196 (rank : 97) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
ZW10_MOUSE
|
||||||
| NC score | 0.048378 (rank : 98) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54692, Q3TIA5, Q3ULW1, Q921H3 | Gene names | Zw10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
MTO1_MOUSE
|
||||||
| NC score | 0.044484 (rank : 99) | θ value | 0.0736092 (rank : 69) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923Z3, Q9D2Q5 | Gene names | Mto1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MTO1 homolog, mitochondrial precursor. | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.036542 (rank : 100) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
IPP4_HUMAN
|
||||||
| NC score | 0.029288 (rank : 101) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.027775 (rank : 102) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.027129 (rank : 103) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.026433 (rank : 104) | θ value | 0.279714 (rank : 79) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.025891 (rank : 105) | θ value | 0.0961366 (rank : 74) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.023699 (rank : 106) | θ value | 0.47712 (rank : 86) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.020148 (rank : 107) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.020125 (rank : 108) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.019775 (rank : 109) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
CR034_HUMAN
|
||||||
| NC score | 0.019451 (rank : 110) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.019270 (rank : 111) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.018825 (rank : 112) | θ value | 0.47712 (rank : 83) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.018379 (rank : 113) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.018277 (rank : 114) | θ value | 0.62314 (rank : 87) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.017445 (rank : 115) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.017298 (rank : 116) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.017173 (rank : 117) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.017166 (rank : 118) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.015912 (rank : 119) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.015684 (rank : 120) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
GCC1_MOUSE
|
||||||
| NC score | 0.015587 (rank : 121) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D4H2, Q8CCR3 | Gene names | Gcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.015252 (rank : 122) | θ value | 1.06291 (rank : 91) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.014992 (rank : 123) | θ value | 0.365318 (rank : 81) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
AN32A_MOUSE
|
||||||
| NC score | 0.014203 (rank : 124) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| NC score | 0.014203 (rank : 125) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
NUB1_HUMAN
|
||||||
| NC score | 0.013985 (rank : 126) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5A7, O95422, Q8IX22, Q9BXR2 | Gene names | NUB1, NYREN18 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD8 ultimate buster 1 (NY-REN-18 antigen). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.013282 (rank : 127) | θ value | 0.47712 (rank : 82) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.013224 (rank : 128) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.013175 (rank : 129) | θ value | 0.279714 (rank : 77) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.013142 (rank : 130) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.012524 (rank : 131) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.011330 (rank : 132) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.011103 (rank : 133) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.010186 (rank : 134) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
ANR52_HUMAN
|
||||||
| NC score | 0.009612 (rank : 135) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| NC score | 0.009576 (rank : 136) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.007925 (rank : 137) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NEBU_HUMAN
|
||||||
| NC score | 0.007538 (rank : 138) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |

|
|||||
| Description | Nebulin. | |||||
|
RP1_MOUSE
|
||||||
| NC score | 0.006513 (rank : 139) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.004061 (rank : 140) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
NBEAL_HUMAN
|
||||||
| NC score | 0.003912 (rank : 141) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS30, Q6Y876, Q6ZQY5, Q96Q30 | Gene names | NBEAL1, ALS2CR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurobeachin-like 1 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.002129 (rank : 142) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ZFP37_MOUSE
|
||||||
| NC score | -0.005283 (rank : 143) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 132 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17141, Q62514 | Gene names | Zfp37, Zfp-37 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 37 (Zfp-37) (Male germ cell-specific zinc finger protein). | |||||