Please be patient as the page loads
|
DDX24_MOUSE
|
||||||
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DDX24_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992652 (rank : 2) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 147 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 2.18568e-46 (rank : 3) | NC score | 0.952062 (rank : 4) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 4.86918e-46 (rank : 4) | NC score | 0.952323 (rank : 3) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 2.58786e-39 (rank : 5) | NC score | 0.945779 (rank : 6) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 1.85459e-37 (rank : 6) | NC score | 0.948169 (rank : 5) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX27_MOUSE
|
||||||
| θ value | 1.24399e-33 (rank : 7) | NC score | 0.937487 (rank : 7) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX18_MOUSE
|
||||||
| θ value | 1.6247e-33 (rank : 8) | NC score | 0.936119 (rank : 8) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX18_HUMAN
|
||||||
| θ value | 2.77131e-33 (rank : 9) | NC score | 0.936024 (rank : 9) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX27_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 10) | NC score | 0.935638 (rank : 11) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 11) | NC score | 0.934685 (rank : 12) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX41_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 12) | NC score | 0.934438 (rank : 14) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| θ value | 1.52067e-31 (rank : 13) | NC score | 0.934093 (rank : 15) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 2.59387e-31 (rank : 14) | NC score | 0.927107 (rank : 25) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 15) | NC score | 0.927242 (rank : 24) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 16) | NC score | 0.934467 (rank : 13) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 1.28732e-30 (rank : 17) | NC score | 0.935995 (rank : 10) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 1.6813e-30 (rank : 18) | NC score | 0.928125 (rank : 20) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 19) | NC score | 0.930346 (rank : 18) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 6.38894e-30 (rank : 20) | NC score | 0.927443 (rank : 23) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX48_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 21) | NC score | 0.922081 (rank : 27) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| θ value | 2.05525e-28 (rank : 22) | NC score | 0.921934 (rank : 28) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 4.57862e-28 (rank : 23) | NC score | 0.909471 (rank : 53) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 4.57862e-28 (rank : 24) | NC score | 0.907059 (rank : 55) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 5.97985e-28 (rank : 25) | NC score | 0.932797 (rank : 16) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 26) | NC score | 0.903142 (rank : 57) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX43_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 27) | NC score | 0.926568 (rank : 26) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX5_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 28) | NC score | 0.920380 (rank : 30) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 29) | NC score | 0.920360 (rank : 31) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX6_MOUSE
|
||||||
| θ value | 4.28545e-26 (rank : 30) | NC score | 0.927720 (rank : 21) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX6_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 31) | NC score | 0.927686 (rank : 22) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX3Y_MOUSE
|
||||||
| θ value | 2.77775e-25 (rank : 32) | NC score | 0.912039 (rank : 49) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 33) | NC score | 0.911035 (rank : 50) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 34) | NC score | 0.928588 (rank : 19) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 8.08199e-25 (rank : 35) | NC score | 0.930556 (rank : 17) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 36) | NC score | 0.910337 (rank : 52) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 37) | NC score | 0.911011 (rank : 51) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX39_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 38) | NC score | 0.918607 (rank : 36) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
IF4A1_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 39) | NC score | 0.916170 (rank : 41) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 40) | NC score | 0.916170 (rank : 42) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A2_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 41) | NC score | 0.915884 (rank : 43) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 42) | NC score | 0.915884 (rank : 44) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX39_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 43) | NC score | 0.919058 (rank : 34) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 44) | NC score | 0.908534 (rank : 54) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
UAP56_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 45) | NC score | 0.917561 (rank : 38) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| θ value | 2.35151e-24 (rank : 46) | NC score | 0.917501 (rank : 39) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX4_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 47) | NC score | 0.918476 (rank : 37) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 1.5242e-23 (rank : 48) | NC score | 0.912741 (rank : 47) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 49) | NC score | 0.912550 (rank : 48) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX53_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 50) | NC score | 0.918840 (rank : 35) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX4_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 51) | NC score | 0.919171 (rank : 33) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX51_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 52) | NC score | 0.916260 (rank : 40) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 53) | NC score | 0.901293 (rank : 58) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 54) | NC score | 0.899593 (rank : 60) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 55) | NC score | 0.893280 (rank : 61) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX1_MOUSE
|
||||||
| θ value | 1.2077e-20 (rank : 56) | NC score | 0.871651 (rank : 67) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX56_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 57) | NC score | 0.920853 (rank : 29) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 58) | NC score | 0.903482 (rank : 56) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX1_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 59) | NC score | 0.870095 (rank : 68) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
DDX56_MOUSE
|
||||||
| θ value | 1.02238e-19 (rank : 60) | NC score | 0.919756 (rank : 32) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 61) | NC score | 0.900362 (rank : 59) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX52_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 62) | NC score | 0.915585 (rank : 45) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DD19A_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 63) | NC score | 0.886691 (rank : 62) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 64) | NC score | 0.885516 (rank : 63) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DDX52_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 65) | NC score | 0.914913 (rank : 46) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DD19A_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 66) | NC score | 0.884956 (rank : 64) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX25_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 67) | NC score | 0.882586 (rank : 66) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 68) | NC score | 0.883068 (rank : 65) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
WRN_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 69) | NC score | 0.470188 (rank : 73) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 70) | NC score | 0.436128 (rank : 74) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 71) | NC score | 0.431007 (rank : 77) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
BLM_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 72) | NC score | 0.476170 (rank : 72) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
BLM_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 73) | NC score | 0.487285 (rank : 71) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
DICER_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 74) | NC score | 0.398745 (rank : 80) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 75) | NC score | 0.397817 (rank : 81) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
RECQ1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 76) | NC score | 0.431140 (rank : 75) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 77) | NC score | 0.529358 (rank : 69) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 78) | NC score | 0.512215 (rank : 70) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 79) | NC score | 0.409796 (rank : 78) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 80) | NC score | 0.406289 (rank : 79) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
HNRPC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 81) | NC score | 0.017555 (rank : 107) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |
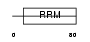
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
MP2K7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 82) | NC score | -0.000790 (rank : 145) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14733, O14648, O14816, O60452, O60453 | Gene names | MAP2K7, JNKK2, MKK7, PRKMK7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity mitogen-activated protein kinase kinase 7 (EC 2.7.12.2) (MAP kinase kinase 7) (MAPKK 7) (MAPK/ERK kinase 7) (JNK-activating kinase 2) (c-Jun N-terminal kinase kinase 2) (JNK kinase 2) (JNKK 2). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 83) | NC score | 0.020309 (rank : 101) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 84) | NC score | 0.014596 (rank : 111) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 85) | NC score | 0.011453 (rank : 115) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 86) | NC score | 0.015916 (rank : 109) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 0.62314 (rank : 87) | NC score | 0.022397 (rank : 98) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
ANC5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.022614 (rank : 97) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTZ4, Q3TGA7, Q80UJ6, Q80W43, Q8BWW7, Q8C0F7, Q9CRX0, Q9CSL1, Q9JJB8 | Gene names | Anapc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | -0.002546 (rank : 148) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
EDAR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.023590 (rank : 96) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNE0, Q9UND9 | Gene names | EDAR, DL | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (EDA- A1 receptor) (Ectodermal dysplasia receptor) (Downless homolog). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.813845 (rank : 91) | NC score | 0.008474 (rank : 119) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 92) | NC score | 0.010002 (rank : 117) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 93) | NC score | 0.006569 (rank : 125) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 94) | NC score | 0.004814 (rank : 130) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
IFIH1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 95) | NC score | 0.301946 (rank : 82) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | -0.003057 (rank : 149) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.015886 (rank : 110) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 98) | NC score | 0.007540 (rank : 120) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RBBP8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 99) | NC score | 0.019284 (rank : 102) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 1.38821 (rank : 100) | NC score | 0.018673 (rank : 103) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
EDAR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.021335 (rank : 99) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.299051 (rank : 83) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
PTN5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | 0.002619 (rank : 136) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
ABL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | -0.005972 (rank : 150) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | -0.001195 (rank : 146) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.000113 (rank : 142) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
IFIH1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.290584 (rank : 84) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | -0.001704 (rank : 147) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.020425 (rank : 100) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
FAM9A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.013806 (rank : 112) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.002846 (rank : 134) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.004674 (rank : 131) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.005132 (rank : 128) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
TE2IP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.012271 (rank : 113) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYB0, Q8WYZ3, Q9NWR2 | Gene names | TERF2IP, RAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1) (hRap1). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.017637 (rank : 106) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CC026_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.139676 (rank : 89) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
CG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.016511 (rank : 108) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.007318 (rank : 121) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
TBX22_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.002755 (rank : 135) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y458, Q96LC0, Q9HBF1 | Gene names | TBX22, TBOX22 | |||
|
Domain Architecture |
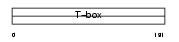
|
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
TRI16_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.006627 (rank : 124) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
ANC5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.017891 (rank : 104) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJX4, Q8N4H7, Q9BQD4 | Gene names | ANAPC5, APC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.017805 (rank : 105) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CING_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.003097 (rank : 133) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.004983 (rank : 129) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
FOXH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.001526 (rank : 140) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88621, Q9QZL5, Q9R241 | Gene names | Foxh1, Fast1, Fast2 | |||
|
Domain Architecture |
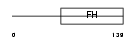
|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.031379 (rank : 95) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.001878 (rank : 139) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
LGP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.290104 (rank : 85) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.033965 (rank : 94) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NR1D2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | -0.000216 (rank : 143) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14995, O00402, Q86XD4 | Gene names | NR1D2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (EAR-1R) (Orphan nuclear hormone receptor BD73). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | -0.000409 (rank : 144) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.431067 (rank : 76) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | -0.007085 (rank : 152) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | -0.006150 (rank : 151) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
U520_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.130630 (rank : 90) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
CO026_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.006916 (rank : 122) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P656, Q8N906 | Gene names | C15orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf26. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.008785 (rank : 118) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.011427 (rank : 116) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
LAP2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.006405 (rank : 127) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
LAP2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.006434 (rank : 126) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.001902 (rank : 138) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.006629 (rank : 123) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.035804 (rank : 93) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.002202 (rank : 137) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
TM131_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.011939 (rank : 114) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.001089 (rank : 141) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.003211 (rank : 132) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CC026_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.154927 (rank : 87) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
DDX58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.151860 (rank : 88) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.183158 (rank : 86) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.088807 (rank : 91) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.069272 (rank : 92) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 147 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.992652 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.952323 (rank : 3) | θ value | 4.86918e-46 (rank : 4) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.952062 (rank : 4) | θ value | 2.18568e-46 (rank : 3) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.948169 (rank : 5) | θ value | 1.85459e-37 (rank : 6) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.945779 (rank : 6) | θ value | 2.58786e-39 (rank : 5) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DDX27_MOUSE
|
||||||
| NC score | 0.937487 (rank : 7) | θ value | 1.24399e-33 (rank : 7) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q921N6, Q8R0W3, Q8R1E2 | Gene names | Ddx27 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX18_MOUSE
|
||||||
| NC score | 0.936119 (rank : 8) | θ value | 1.6247e-33 (rank : 8) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8K363, Q3MIB0, Q8BVZ2, Q9D2E0 | Gene names | Ddx18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18). | |||||
|
DDX18_HUMAN
|
||||||
| NC score | 0.936024 (rank : 9) | θ value | 2.77131e-33 (rank : 9) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9NVP1, Q6IAU4, Q92732, Q9BQB7 | Gene names | DDX18 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX18 (EC 3.6.1.-) (DEAD box protein 18) (Myc-regulated DEAD box protein) (MrDb). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.935995 (rank : 10) | θ value | 1.28732e-30 (rank : 17) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX27_HUMAN
|
||||||
| NC score | 0.935638 (rank : 11) | θ value | 8.06329e-33 (rank : 10) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q96GQ7, Q5VXM7, Q8WYG4, Q969N7, Q96F57, Q96L97, Q9BWY9, Q9BXF0, Q9H990, Q9NWU3, Q9P0C2, Q9UGD6 | Gene names | DDX27, RHLP | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX27 (EC 3.6.1.-) (DEAD box protein 27). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.934685 (rank : 12) | θ value | 5.22648e-32 (rank : 11) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.934467 (rank : 13) | θ value | 9.8567e-31 (rank : 16) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
DDX41_HUMAN
|
||||||
| NC score | 0.934438 (rank : 14) | θ value | 1.52067e-31 (rank : 12) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9UJV9, Q96BK6, Q96K05, Q9NT96, Q9NW04 | Gene names | DDX41, ABS | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41) (DEAD box protein abstrakt homolog). | |||||
|
DDX41_MOUSE
|
||||||
| NC score | 0.934093 (rank : 15) | θ value | 1.52067e-31 (rank : 13) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q91VN6 | Gene names | Ddx41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX41 (EC 3.6.1.-) (DEAD box protein 41). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.932797 (rank : 16) | θ value | 5.97985e-28 (rank : 25) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.930556 (rank : 17) | θ value | 8.08199e-25 (rank : 35) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.930346 (rank : 18) | θ value | 3.74554e-30 (rank : 19) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.928588 (rank : 19) | θ value | 6.18819e-25 (rank : 34) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.928125 (rank : 20) | θ value | 1.6813e-30 (rank : 18) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX6_MOUSE
|
||||||
| NC score | 0.927720 (rank : 21) | θ value | 4.28545e-26 (rank : 30) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P54823 | Gene names | Ddx6, Hlr2, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK homolog). | |||||
|
DDX6_HUMAN
|
||||||
| NC score | 0.927686 (rank : 22) | θ value | 2.12685e-25 (rank : 31) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P26196 | Gene names | DDX6, HLR2, RCK | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX6 (EC 3.6.1.-) (DEAD box protein 6) (ATP-dependent RNA helicase p54) (Oncogene RCK). | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.927443 (rank : 23) | θ value | 6.38894e-30 (rank : 20) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.927242 (rank : 24) | θ value | 2.59387e-31 (rank : 15) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.927107 (rank : 25) | θ value | 2.59387e-31 (rank : 14) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
DDX43_HUMAN
|
||||||
| NC score | 0.926568 (rank : 26) | θ value | 3.87602e-27 (rank : 27) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9NXZ2, Q6NXR1 | Gene names | DDX43, HAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX43 (EC 3.6.1.-) (DEAD box protein 43) (DEAD box protein HAGE) (Helical antigen). | |||||
|
DDX48_HUMAN
|
||||||
| NC score | 0.922081 (rank : 27) | θ value | 2.05525e-28 (rank : 21) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P38919, Q15033, Q96A18 | Gene names | DDX48, EIF4A3, KIAA0111 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48) (Eukaryotic initiation factor 4A-like NUK-34) (Nuclear matrix protein 265) (hNMP 265) (Eukaryotic translation initiation factor 4A isoform 3). | |||||
|
DDX48_MOUSE
|
||||||
| NC score | 0.921934 (rank : 28) | θ value | 2.05525e-28 (rank : 22) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q91VC3 | Gene names | Ddx48 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX48 (EC 3.6.1.-) (DEAD box protein 48). | |||||
|
DDX56_HUMAN
|
||||||
| NC score | 0.920853 (rank : 29) | θ value | 1.2077e-20 (rank : 57) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NY93, Q9H9I8 | Gene names | DDX56, DDX21, NOH61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase) (DEAD-box protein 21). | |||||
|
DDX5_HUMAN
|
||||||
| NC score | 0.920380 (rank : 30) | θ value | 1.12775e-26 (rank : 28) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P17844, O75681 | Gene names | DDX5, HELR, HLR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68). | |||||
|
DDX5_MOUSE
|
||||||
| NC score | 0.920360 (rank : 31) | θ value | 1.12775e-26 (rank : 29) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q61656 | Gene names | Ddx5, Tnz2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX5 (EC 3.6.1.-) (DEAD box protein 5) (RNA helicase p68) (DEAD box RNA helicase DEAD1) (mDEAD1). | |||||
|
DDX56_MOUSE
|
||||||
| NC score | 0.919756 (rank : 32) | θ value | 1.02238e-19 (rank : 60) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9D0R4 | Gene names | Ddx56, D11Ertd619e, Noh61 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX56 (EC 3.6.1.-) (DEAD box protein 56) (ATP-dependent 61 kDa nucleolar RNA helicase). | |||||
|
DDX4_MOUSE
|
||||||
| NC score | 0.919171 (rank : 33) | θ value | 7.56453e-23 (rank : 51) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q61496, Q9D5X7 | Gene names | Ddx4, Vasa | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog) (Mvh). | |||||
|
DDX39_HUMAN
|
||||||
| NC score | 0.919058 (rank : 34) | θ value | 2.35151e-24 (rank : 43) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O00148, Q9BVP6, Q9H5W0 | Gene names | DDX39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39) (Nuclear RNA helicase URH49). | |||||
|
DDX53_HUMAN
|
||||||
| NC score | 0.918840 (rank : 35) | θ value | 4.43474e-23 (rank : 50) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q86TM3, Q6NVV4 | Gene names | DDX53, CAGE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX53 (EC 3.6.1.-) (DEAD box protein 53) (DEAD box protein CAGE) (Cancer-associated gene protein). | |||||
|
DDX39_MOUSE
|
||||||
| NC score | 0.918607 (rank : 36) | θ value | 1.37858e-24 (rank : 38) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8VDW0, Q3UJV4, Q8C2C2 | Gene names | Ddx39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX39 (EC 3.6.1.-) (DEAD box protein 39). | |||||
|
DDX4_HUMAN
|
||||||
| NC score | 0.918476 (rank : 37) | θ value | 1.5242e-23 (rank : 47) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9NQI0, Q86VX0, Q9NT92, Q9NYB1 | Gene names | DDX4, VASA | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX4 (EC 3.6.1.-) (DEAD box protein 4) (VASA homolog). | |||||
|
UAP56_HUMAN
|
||||||
| NC score | 0.917561 (rank : 38) | θ value | 2.35151e-24 (rank : 45) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q13838, O43496, Q5RJ64, Q5RJ66, Q5ST94, Q5STB4, Q5STB5, Q5STB7, Q5STU4, Q5STU5, Q5STU6, Q5STU8, Q71V76 | Gene names | BAT1, UAP56 | |||
|
Domain Architecture |

|
|||||
| Description | Spliceosome RNA helicase BAT1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (ATP-dependent RNA helicase p47) (HLA-B-associated transcript-1). | |||||
|
UAP56_MOUSE
|
||||||
| NC score | 0.917501 (rank : 39) | θ value | 2.35151e-24 (rank : 46) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9Z1N5, Q8HW97 | Gene names | Bat1, Bat1a, Uap56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spliceosome RNA helicase Bat1 (EC 3.6.1.-) (DEAD box protein UAP56) (56 kDa U2AF65-associated protein) (HLA-B-associated transcript 1). | |||||
|
DDX51_MOUSE
|
||||||
| NC score | 0.916260 (rank : 40) | θ value | 1.6852e-22 (rank : 52) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q6P9R1, Q3U7M2 | Gene names | Ddx51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
IF4A1_HUMAN
|
||||||
| NC score | 0.916170 (rank : 41) | θ value | 1.37858e-24 (rank : 39) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P60842, P04765, Q5U018, Q61516 | Gene names | EIF4A1, DDX2A, EIF4A | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A1_MOUSE
|
||||||
| NC score | 0.916170 (rank : 42) | θ value | 1.37858e-24 (rank : 40) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P60843, P04765, Q61516 | Gene names | Eif4a1, Ddx2a, Eif4a | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-I (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-1) (eIF4A-I) (eIF-4A-I). | |||||
|
IF4A2_HUMAN
|
||||||
| NC score | 0.915884 (rank : 43) | θ value | 1.37858e-24 (rank : 41) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14240, Q96B90, Q96EA8 | Gene names | EIF4A2, DDX2B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
IF4A2_MOUSE
|
||||||
| NC score | 0.915884 (rank : 44) | θ value | 1.37858e-24 (rank : 42) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P10630, Q61513, Q61514 | Gene names | Eif4a2, Ddx2b | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic initiation factor 4A-II (EC 3.6.1.-) (ATP-dependent RNA helicase eIF4A-2) (eIF4A-II) (eIF-4A-II). | |||||
|
DDX52_HUMAN
|
||||||
| NC score | 0.915585 (rank : 45) | θ value | 1.47631e-18 (rank : 62) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Y2R4, Q86YG1, Q8N213, Q9NVE0, Q9Y482 | Gene names | DDX52, ROK1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX52_MOUSE
|
||||||
| NC score | 0.914913 (rank : 46) | θ value | 6.20254e-17 (rank : 65) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q8K301, Q8BV29 | Gene names | Ddx52, Rok1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX52 (EC 3.6.1.-) (DEAD box protein 52) (ATP-dependent RNA helicase ROK1-like). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.912741 (rank : 47) | θ value | 1.5242e-23 (rank : 48) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.912550 (rank : 48) | θ value | 1.99067e-23 (rank : 49) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX3Y_MOUSE
|
||||||
| NC score | 0.912039 (rank : 49) | θ value | 2.77775e-25 (rank : 32) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q62095, Q9QWS9 | Gene names | Ddx3y, D1Pas1-rs1, Dead2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal) (DEAD-box RNA helicase DEAD2) (mDEAD2) (D1Pas1-related sequence 1). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.911035 (rank : 50) | θ value | 4.73814e-25 (rank : 33) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.911011 (rank : 51) | θ value | 1.05554e-24 (rank : 37) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.910337 (rank : 52) | θ value | 1.05554e-24 (rank : 36) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.909471 (rank : 53) | θ value | 4.57862e-28 (rank : 23) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.908534 (rank : 54) | θ value | 2.35151e-24 (rank : 44) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.907059 (rank : 55) | θ value | 4.57862e-28 (rank : 24) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.903482 (rank : 56) | θ value | 5.99374e-20 (rank : 58) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.903142 (rank : 57) | θ value | 2.27234e-27 (rank : 26) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.901293 (rank : 58) | θ value | 2.87452e-22 (rank : 53) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.900362 (rank : 59) | θ value | 1.33526e-19 (rank : 61) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.899593 (rank : 60) | θ value | 1.86321e-21 (rank : 54) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.893280 (rank : 61) | θ value | 4.15078e-21 (rank : 55) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DD19A_HUMAN
|
||||||
| NC score | 0.886691 (rank : 62) | θ value | 1.24977e-17 (rank : 63) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9NUU7, Q53FM0 | Gene names | DDX19A, DDX19L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DDX19-like protein). | |||||
|
DD19B_HUMAN
|
||||||
| NC score | 0.885516 (rank : 63) | θ value | 3.63628e-17 (rank : 64) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UMR2, Q6FIB7, Q6IAE0 | Gene names | DDX19B, DBP5, DDX19 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19B (EC 3.6.1.-) (DEAD box protein 19B) (DEAD box RNA helicase DEAD5). | |||||
|
DD19A_MOUSE
|
||||||
| NC score | 0.884956 (rank : 64) | θ value | 1.80466e-16 (rank : 66) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61655, Q921R0 | Gene names | Ddx19a, Ddx19, Eif4a-rs1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX19A (EC 3.6.1.-) (DEAD box protein 19A) (DEAD box RNA helicase DEAD5) (mDEAD5) (Eukaryotic translation initiation factor 4A-related sequence 1). | |||||
|
DDX25_HUMAN
|
||||||
| NC score | 0.883068 (rank : 65) | θ value | 1.09485e-13 (rank : 68) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UHL0, Q86W81, Q8IYP1 | Gene names | DDX25, GRTH | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX25_MOUSE
|
||||||
| NC score | 0.882586 (rank : 66) | θ value | 3.76295e-14 (rank : 67) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QY15, Q8R1B6 | Gene names | Ddx25, Grth | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX25 (EC 3.6.1.-) (DEAD box protein 25) (Gonadotropin-regulated testicular RNA helicase). | |||||
|
DDX1_MOUSE
|
||||||
| NC score | 0.871651 (rank : 67) | θ value | 1.2077e-20 (rank : 56) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q91VR5, Q3TU41 | Gene names | Ddx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1). | |||||
|
DDX1_HUMAN
|
||||||
| NC score | 0.870095 (rank : 68) | θ value | 7.82807e-20 (rank : 59) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q92499 | Gene names | DDX1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX1 (EC 3.6.1.-) (DEAD box protein 1) (DEAD box protein retinoblastoma) (DBP-RB). | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.529358 (rank : 69) | θ value | 0.00869519 (rank : 77) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.512215 (rank : 70) | θ value | 0.0148317 (rank : 78) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
BLM_MOUSE
|
||||||
| NC score | 0.487285 (rank : 71) | θ value | 0.00665767 (rank : 73) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O88700, O88198 | Gene names | Blm | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein homolog (EC 3.6.1.-) (mBLM). | |||||
|
BLM_HUMAN
|
||||||
| NC score | 0.476170 (rank : 72) | θ value | 0.00390308 (rank : 72) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P54132 | Gene names | BLM, RECQ2, RECQL3 | |||
|
Domain Architecture |

|
|||||
| Description | Bloom syndrome protein (EC 3.6.1.-) (RecQ protein-like 3) (DNA helicase, RecQ-like type 2). | |||||
|
WRN_HUMAN
|
||||||
| NC score | 0.470188 (rank : 73) | θ value | 2.44474e-05 (rank : 69) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q14191 | Gene names | WRN, RECQ3, RECQL2 | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase (EC 3.6.1.-). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.436128 (rank : 74) | θ value | 9.29e-05 (rank : 70) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ1_HUMAN
|
||||||
| NC score | 0.431140 (rank : 75) | θ value | 0.00869519 (rank : 76) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P46063 | Gene names | RECQL, RECQL1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.431067 (rank : 76) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.431007 (rank : 77) | θ value | 0.00134147 (rank : 71) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.409796 (rank : 78) | θ value | 0.0330416 (rank : 79) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FANCM_MOUSE
|
||||||
| NC score | 0.406289 (rank : 79) | θ value | 0.0563607 (rank : 80) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8BGE5, Q69ZF5, Q8BKB7, Q8BUA8 | Gene names | Fancm, Kiaa1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM). | |||||
|
DICER_HUMAN
|
||||||
| NC score | 0.398745 (rank : 80) | θ value | 0.00665767 (rank : 74) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UPY3, O95943, Q9UQ02 | Gene names | DICER1, DICER, HERNA, KIAA0928 | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Helicase with RNase motif) (Helicase-MOI). | |||||
|
DICER_MOUSE
|
||||||
| NC score | 0.397817 (rank : 81) | θ value | 0.00665767 (rank : 75) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R418 | Gene names | Dicer1, Dicer, Mdcr | |||
|
Domain Architecture |

|
|||||
| Description | Endoribonuclease Dicer (EC 3.1.26.-) (Double-strand-specific ribonuclease mDCR-1). | |||||
|
IFIH1_HUMAN
|
||||||
| NC score | 0.301946 (rank : 82) | θ value | 1.06291 (rank : 95) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9BYX4, Q6DC96, Q86X56, Q96MX8, Q9H3G6 | Gene names | IFIH1, MDA5, RH116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon-induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5) (RNA helicase-DEAD box protein 116) (Murabutide-down- regulated protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.299051 (rank : 83) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
IFIH1_MOUSE
|
||||||
| NC score | 0.290584 (rank : 84) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8R5F7, Q3U6S2, Q68EM4, Q8BYC9, Q8BZ01, Q8K5C7, Q8R144, Q8VE79, Q99KS4, Q9D2Z5 | Gene names | Ifih1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-induced helicase C domain-containing protein 1 (EC 3.6.1.-) (Interferon induced with helicase C domain protein 1) (Helicase with 2 CARD domains) (Helicard) (Melanoma differentiation-associated protein 5) (MDA-5). | |||||
|
LGP2_HUMAN
|
||||||
| NC score | 0.290104 (rank : 85) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96C10, Q9HAM6 | Gene names | LGP2, D11LGP2E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2 homolog). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.183158 (rank : 86) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
CC026_HUMAN
|
||||||
| NC score | 0.154927 (rank : 87) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ75 | Gene names | C3orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26. | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.151860 (rank : 88) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
CC026_MOUSE
|
||||||
| NC score | 0.139676 (rank : 89) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CZT6, Q9CY77 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf26 homolog. | |||||
|
U520_HUMAN
|
||||||
| NC score | 0.130630 (rank : 90) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75643, O94884, Q6PX59, Q96IF2, Q9H7S0 | Gene names | ASCC3L1, HELIC2, KIAA0788 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U5 small nuclear ribonucleoprotein 200 kDa helicase (EC 3.6.1.-) (U5 snRNP-specific 200 kDa protein) (U5-200KD) (Activating signal cointegrator 1 complex subunit 3-like 1) (BRR2 homolog). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.088807 (rank : 91) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.069272 (rank : 92) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.035804 (rank : 93) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.033965 (rank : 94) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.031379 (rank : 95) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
EDAR_HUMAN
|
||||||
| NC score | 0.023590 (rank : 96) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UNE0, Q9UND9 | Gene names | EDAR, DL | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (EDA- A1 receptor) (Ectodermal dysplasia receptor) (Downless homolog). | |||||
|
ANC5_MOUSE
|
||||||
| NC score | 0.022614 (rank : 97) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTZ4, Q3TGA7, Q80UJ6, Q80W43, Q8BWW7, Q8C0F7, Q9CRX0, Q9CSL1, Q9JJB8 | Gene names | Anapc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.022397 (rank : 98) | θ value | 0.62314 (rank : 87) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
EDAR_MOUSE
|
||||||
| NC score | 0.021335 (rank : 99) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.020425 (rank : 100) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.020309 (rank : 101) | θ value | 0.21417 (rank : 83) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
RBBP8_HUMAN
|
||||||
| NC score | 0.019284 (rank : 102) | θ value | 1.38821 (rank : 99) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.018673 (rank : 103) | θ value | 1.38821 (rank : 100) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
ANC5_HUMAN
|
||||||
| NC score | 0.017891 (rank : 104) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJX4, Q8N4H7, Q9BQD4 | Gene names | ANAPC5, APC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.017805 (rank : 105) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.017637 (rank : 106) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
HNRPC_MOUSE
|
||||||
| NC score | 0.017555 (rank : 107) | θ value | 0.163984 (rank : 81) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.016511 (rank : 108) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.015916 (rank : 109) | θ value | 0.62314 (rank : 86) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.015886 (rank : 110) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.014596 (rank : 111) | θ value | 0.279714 (rank : 84) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
FAM9A_HUMAN
|
||||||
| NC score | 0.013806 (rank : 112) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
TE2IP_HUMAN
|
||||||
| NC score | 0.012271 (rank : 113) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYB0, Q8WYZ3, Q9NWR2 | Gene names | TERF2IP, RAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomeric repeat-binding factor 2-interacting protein 1 (TRF2- interacting telomeric protein Rap1) (hRap1). | |||||
|
TM131_MOUSE
|
||||||
| NC score | 0.011939 (rank : 114) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.011453 (rank : 115) | θ value | 0.62314 (rank : 85) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.011427 (rank : 116) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.010002 (rank : 117) | θ value | 1.06291 (rank : 92) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.008785 (rank : 118) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.008474 (rank : 119) | θ value | 0.813845 (rank : 91) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.007540 (rank : 120) | θ value | 1.38821 (rank : 98) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.007318 (rank : 121) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
CO026_HUMAN
|
||||||
| NC score | 0.006916 (rank : 122) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P656, Q8N906 | Gene names | C15orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf26. | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.006629 (rank : 123) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TRI16_HUMAN
|
||||||
| NC score | 0.006627 (rank : 124) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95361, Q7Z6I2, Q96BE8, Q96J43 | Gene names | TRIM16, EBBP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 16 (Estrogen-responsive B box protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.006569 (rank : 125) | θ value | 1.06291 (rank : 93) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
LAP2B_MOUSE
|
||||||
| NC score | 0.006434 (rank : 126) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
|
LAP2A_MOUSE
|
||||||
| NC score | 0.006405 (rank : 127) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.005132 (rank : 128) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.004983 (rank : 129) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.004814 (rank : 130) | θ value | 1.06291 (rank : 94) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.004674 (rank : 131) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.003211 (rank : 132) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.003097 (rank : 133) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.002846 (rank : 134) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
TBX22_HUMAN
|
||||||
| NC score | 0.002755 (rank : 135) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y458, Q96LC0, Q9HBF1 | Gene names | TBX22, TBOX22 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX22 (T-box protein 22). | |||||
|
PTN5_MOUSE
|
||||||
| NC score | 0.002619 (rank : 136) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54830, Q64694 | Gene names | Ptpn5 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 5 (EC 3.1.3.48) (Protein-tyrosine phosphatase striatum-enriched) (STEP) (Neural- specific protein-tyrosine phosphatase). | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.002202 (rank : 137) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.001902 (rank : 138) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.001878 (rank : 139) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
FOXH1_MOUSE
|
||||||
| NC score | 0.001526 (rank : 140) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88621, Q9QZL5, Q9R241 | Gene names | Foxh1, Fast1, Fast2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.001089 (rank : 141) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.000113 (rank : 142) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
NR1D2_HUMAN
|
||||||
| NC score | -0.000216 (rank : 143) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14995, O00402, Q86XD4 | Gene names | NR1D2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (EAR-1R) (Orphan nuclear hormone receptor BD73). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | -0.000409 (rank : 144) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
MP2K7_HUMAN
|
||||||
| NC score | -0.000790 (rank : 145) | θ value | 0.21417 (rank : 82) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 833 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14733, O14648, O14816, O60452, O60453 | Gene names | MAP2K7, JNKK2, MKK7, PRKMK7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity mitogen-activated protein kinase kinase 7 (EC 2.7.12.2) (MAP kinase kinase 7) (MAPKK 7) (MAPK/ERK kinase 7) (JNK-activating kinase 2) (c-Jun N-terminal kinase kinase 2) (JNK kinase 2) (JNKK 2). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | -0.001195 (rank : 146) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | -0.001704 (rank : 147) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | -0.002546 (rank : 148) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | -0.003057 (rank : 149) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ABL2_HUMAN
|
||||||
| NC score | -0.005972 (rank : 150) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1045 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42684, Q5T0X6, Q6NZY6 | Gene names | ABL2, ABLL, ARG | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase ABL2 (EC 2.7.10.2) (Abelson murine leukemia viral oncogene homolog 2) (Tyrosine kinase ARG). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | -0.006150 (rank : 151) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.007085 (rank : 152) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 147 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||