Please be patient as the page loads
|
LAP2B_MOUSE
|
||||||
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LAP2B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.988552 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42167, Q14861 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
LAP2B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
|
LAP2A_MOUSE
|
||||||
| θ value | 6.95003e-85 (rank : 3) | NC score | 0.925510 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
LAP2A_HUMAN
|
||||||
| θ value | 7.44272e-79 (rank : 4) | NC score | 0.924101 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42166, P08918, P08919, Q14860, Q16295 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
EMD_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 5) | NC score | 0.300892 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50402 | Gene names | EMD, EDMD, STA | |||
|
Domain Architecture |

|
|||||
| Description | Emerin. | |||||
|
EMD_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 6) | NC score | 0.307702 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08579 | Gene names | Emd, Sta | |||
|
Domain Architecture |

|
|||||
| Description | Emerin. | |||||
|
MAN1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.187220 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2U8, Q9NT47, Q9NYA5 | Gene names | LEMD3, MAN1 | |||
|
Domain Architecture |
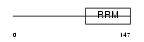
|
|||||
| Description | Inner nuclear membrane protein Man1 (LEM domain-containing protein 3). | |||||
|
CCNI_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.113602 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.063359 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.015584 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
AHTF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.034482 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.063569 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.036025 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AT2C2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.025653 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.052746 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.043901 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.002274 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ZFP37_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.004892 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6Q3 | Gene names | ZFP37 | |||
|
Domain Architecture |
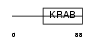
|
|||||
| Description | Zinc finger protein 37 homolog (Zfp-37). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.029536 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.074249 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.021151 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
GALT5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.008064 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
CCNI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.066311 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |
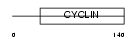
|
|||||
| Description | Cyclin-I. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.016938 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.016946 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.010690 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.007744 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
DDX24_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.006434 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
LAP2B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
|
LAP2B_HUMAN
|
||||||
| NC score | 0.988552 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P42167, Q14861 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
LAP2A_MOUSE
|
||||||
| NC score | 0.925510 (rank : 3) | θ value | 6.95003e-85 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
LAP2A_HUMAN
|
||||||
| NC score | 0.924101 (rank : 4) | θ value | 7.44272e-79 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42166, P08918, P08919, Q14860, Q16295 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
EMD_MOUSE
|
||||||
| NC score | 0.307702 (rank : 5) | θ value | 0.00134147 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08579 | Gene names | Emd, Sta | |||
|
Domain Architecture |

|
|||||
| Description | Emerin. | |||||
|
EMD_HUMAN
|
||||||
| NC score | 0.300892 (rank : 6) | θ value | 0.00134147 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50402 | Gene names | EMD, EDMD, STA | |||
|
Domain Architecture |

|
|||||
| Description | Emerin. | |||||
|
MAN1_HUMAN
|
||||||
| NC score | 0.187220 (rank : 7) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2U8, Q9NT47, Q9NYA5 | Gene names | LEMD3, MAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Inner nuclear membrane protein Man1 (LEM domain-containing protein 3). | |||||
|
CCNI_HUMAN
|
||||||
| NC score | 0.113602 (rank : 8) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.074249 (rank : 9) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
CCNI_MOUSE
|
||||||
| NC score | 0.066311 (rank : 10) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.063569 (rank : 11) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.063359 (rank : 12) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.052746 (rank : 13) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.043901 (rank : 14) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.036025 (rank : 15) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
AHTF1_MOUSE
|
||||||
| NC score | 0.034482 (rank : 16) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.029536 (rank : 17) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
AT2C2_HUMAN
|
||||||
| NC score | 0.025653 (rank : 18) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75185 | Gene names | KIAA0703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable calcium-transporting ATPase KIAA0703 (EC 3.6.3.8). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.021151 (rank : 19) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.016946 (rank : 20) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.016938 (rank : 21) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.015584 (rank : 22) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.010690 (rank : 23) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
GALT5_HUMAN
|
||||||
| NC score | 0.008064 (rank : 24) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.007744 (rank : 25) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
DDX24_MOUSE
|
||||||
| NC score | 0.006434 (rank : 26) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESV0, Q61119 | Gene names | Ddx24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
ZFP37_HUMAN
|
||||||
| NC score | 0.004892 (rank : 27) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 797 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6Q3 | Gene names | ZFP37 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 37 homolog (Zfp-37). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.002274 (rank : 28) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||