Please be patient as the page loads
|
CCNI_HUMAN
|
||||||
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNI_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994734 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |
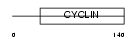
|
|||||
| Description | Cyclin-I. | |||||
|
CCNG2_HUMAN
|
||||||
| θ value | 4.57862e-28 (rank : 3) | NC score | 0.807022 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |
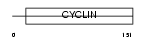
|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_MOUSE
|
||||||
| θ value | 1.33217e-27 (rank : 4) | NC score | 0.807396 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |
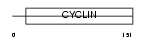
|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG1_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 5) | NC score | 0.790380 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |
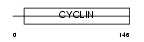
|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 6) | NC score | 0.786748 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNE2_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 7) | NC score | 0.638109 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |
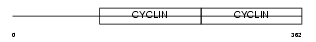
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE2_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 8) | NC score | 0.628174 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |
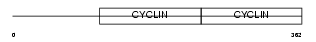
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 9) | NC score | 0.609057 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |
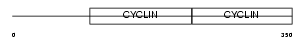
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCND2_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 10) | NC score | 0.584721 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |
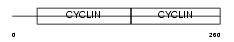
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCNE1_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 11) | NC score | 0.589007 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCND2_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 12) | NC score | 0.583934 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 13) | NC score | 0.593698 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCND1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 14) | NC score | 0.588217 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCNB2_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 15) | NC score | 0.534869 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
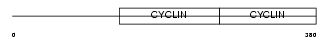
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 16) | NC score | 0.542458 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 17) | NC score | 0.541358 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNB2_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 18) | NC score | 0.526843 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
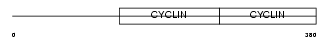
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 19) | NC score | 0.506582 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
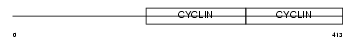
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 20) | NC score | 0.504835 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
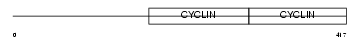
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 21) | NC score | 0.395207 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCND3_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 22) | NC score | 0.550675 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |
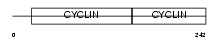
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 23) | NC score | 0.518178 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 24) | NC score | 0.526133 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
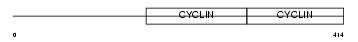
|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 25) | NC score | 0.458993 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCND3_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 26) | NC score | 0.538596 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |
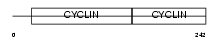
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
LAP2A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 27) | NC score | 0.123646 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42166, P08918, P08919, Q14860, Q16295 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
LAP2B_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 28) | NC score | 0.125820 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42167, Q14861 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
LAP2A_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.110410 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
LAP2B_MOUSE
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.113602 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
|
UNG2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 31) | NC score | 0.242582 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |
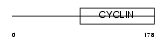
|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.111056 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.089994 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
PSMD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.018566 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99460, Q53TI2, Q6GMU5, Q6P2P4, Q6PJM7, Q6PKG9, Q86VU1, Q8IV79 | Gene names | PSMD1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 1 (26S proteasome regulatory subunit RPN2) (26S proteasome regulatory subunit S1) (26S proteasome subunit p112). | |||||
|
CPSF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.031339 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
FASTK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.017904 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIX9 | Gene names | Fastk | |||
|
Domain Architecture |

|
|||||
| Description | Fas-activated serine/threonine kinase (EC 2.7.11.8) (FAST kinase). | |||||
|
CPSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.029384 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.008179 (rank : 40) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MMP13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.002940 (rank : 41) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
CCNF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.254340 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.252339 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNI_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_MOUSE
|
||||||
| NC score | 0.994734 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNG2_MOUSE
|
||||||
| NC score | 0.807396 (rank : 3) | θ value | 1.33217e-27 (rank : 4) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_HUMAN
|
||||||
| NC score | 0.807022 (rank : 4) | θ value | 4.57862e-28 (rank : 3) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG1_HUMAN
|
||||||
| NC score | 0.790380 (rank : 5) | θ value | 5.79196e-23 (rank : 5) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_MOUSE
|
||||||
| NC score | 0.786748 (rank : 6) | θ value | 1.09232e-21 (rank : 6) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNE2_HUMAN
|
||||||
| NC score | 0.638109 (rank : 7) | θ value | 4.16044e-13 (rank : 7) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE2_MOUSE
|
||||||
| NC score | 0.628174 (rank : 8) | θ value | 1.58096e-12 (rank : 8) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_HUMAN
|
||||||
| NC score | 0.609057 (rank : 9) | θ value | 6.64225e-11 (rank : 9) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCND1_HUMAN
|
||||||
| NC score | 0.593698 (rank : 10) | θ value | 1.80886e-08 (rank : 13) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCNE1_MOUSE
|
||||||
| NC score | 0.589007 (rank : 11) | θ value | 8.11959e-09 (rank : 11) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCND1_MOUSE
|
||||||
| NC score | 0.588217 (rank : 12) | θ value | 3.08544e-08 (rank : 14) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCND2_HUMAN
|
||||||
| NC score | 0.584721 (rank : 13) | θ value | 8.11959e-09 (rank : 10) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND2_MOUSE
|
||||||
| NC score | 0.583934 (rank : 14) | θ value | 1.06045e-08 (rank : 12) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND3_HUMAN
|
||||||
| NC score | 0.550675 (rank : 15) | θ value | 4.1701e-05 (rank : 22) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.542458 (rank : 16) | θ value | 6.43352e-06 (rank : 16) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.541358 (rank : 17) | θ value | 6.43352e-06 (rank : 17) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCND3_MOUSE
|
||||||
| NC score | 0.538596 (rank : 18) | θ value | 0.000270298 (rank : 26) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNB2_HUMAN
|
||||||
| NC score | 0.534869 (rank : 19) | θ value | 4.92598e-06 (rank : 15) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB2_MOUSE
|
||||||
| NC score | 0.526843 (rank : 20) | θ value | 1.09739e-05 (rank : 18) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.526133 (rank : 21) | θ value | 9.29e-05 (rank : 24) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.518178 (rank : 22) | θ value | 9.29e-05 (rank : 23) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB1_MOUSE
|
||||||
| NC score | 0.506582 (rank : 23) | θ value | 1.43324e-05 (rank : 19) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_HUMAN
|
||||||
| NC score | 0.504835 (rank : 24) | θ value | 1.87187e-05 (rank : 20) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.458993 (rank : 25) | θ value | 0.000158464 (rank : 25) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.395207 (rank : 26) | θ value | 1.87187e-05 (rank : 21) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNF_HUMAN
|
||||||
| NC score | 0.254340 (rank : 27) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_MOUSE
|
||||||
| NC score | 0.252339 (rank : 28) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
UNG2_HUMAN
|
||||||
| NC score | 0.242582 (rank : 29) | θ value | 0.125558 (rank : 31) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
LAP2B_HUMAN
|
||||||
| NC score | 0.125820 (rank : 30) | θ value | 0.0330416 (rank : 28) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42167, Q14861 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2, isoforms beta/gamma (Thymopoietin, isoforms beta/gamma) (TP beta/gamma) (Thymopoietin-related peptide isoforms beta/gamma) (TPRP isoforms beta/gamma) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
LAP2A_HUMAN
|
||||||
| NC score | 0.123646 (rank : 31) | θ value | 0.0330416 (rank : 27) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42166, P08918, P08919, Q14860, Q16295 | Gene names | TMPO, LAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoform alpha (Thymopoietin isoform alpha) (TP alpha) (Thymopoietin-related peptide isoform alpha) (TPRP isoform alpha) [Contains: Thymopoietin (TP) (Splenin); Thymopentin (TP5)]. | |||||
|
LAP2B_MOUSE
|
||||||
| NC score | 0.113602 (rank : 32) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61029, Q61030, Q61031, Q61032 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms beta/delta/epsilon/gamma (Thymopoietin isoforms beta/delta/epsilon/gamma) (TP beta/delta/epsilon/gamma). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.111056 (rank : 33) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
LAP2A_MOUSE
|
||||||
| NC score | 0.110410 (rank : 34) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61033, Q61028 | Gene names | Tmpo, Lap2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamina-associated polypeptide 2 isoforms alpha/zeta (Thymopoietin isoforms alpha/zeta) (TP alpha/zeta). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.089994 (rank : 35) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CPSF1_HUMAN
|
||||||
| NC score | 0.031339 (rank : 36) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
CPSF1_MOUSE
|
||||||
| NC score | 0.029384 (rank : 37) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
PSMD1_HUMAN
|
||||||
| NC score | 0.018566 (rank : 38) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99460, Q53TI2, Q6GMU5, Q6P2P4, Q6PJM7, Q6PKG9, Q86VU1, Q8IV79 | Gene names | PSMD1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 1 (26S proteasome regulatory subunit RPN2) (26S proteasome regulatory subunit S1) (26S proteasome subunit p112). | |||||
|
FASTK_MOUSE
|
||||||
| NC score | 0.017904 (rank : 39) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIX9 | Gene names | Fastk | |||
|
Domain Architecture |

|
|||||
| Description | Fas-activated serine/threonine kinase (EC 2.7.11.8) (FAST kinase). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.008179 (rank : 40) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MMP13_HUMAN
|
||||||
| NC score | 0.002940 (rank : 41) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 39 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||