Please be patient as the page loads
|
CCNF_MOUSE
|
||||||
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNF_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992573 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 3) | NC score | 0.731710 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 4) | NC score | 0.730050 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 5) | NC score | 0.727456 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | 1.74391e-19 (rank : 6) | NC score | 0.720649 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 7) | NC score | 0.663048 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNB1_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 8) | NC score | 0.702533 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 9) | NC score | 0.693354 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB1_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 10) | NC score | 0.701730 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 11) | NC score | 0.690962 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |
|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 12) | NC score | 0.564845 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
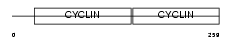
CCND1_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 13) | NC score | 0.541806 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
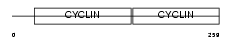
CCND1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 14) | NC score | 0.543318 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
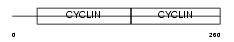
CCND2_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 15) | NC score | 0.520542 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
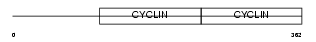
CCNE2_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 16) | NC score | 0.542038 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
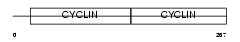
CCND2_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 17) | NC score | 0.506121 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
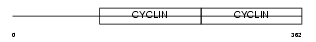
CCNE2_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 18) | NC score | 0.538753 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
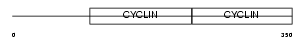
CCNE1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 19) | NC score | 0.523120 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCNE1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 20) | NC score | 0.516113 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
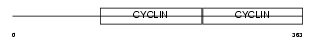
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCND3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 21) | NC score | 0.489197 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
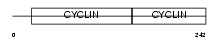
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND3_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 22) | NC score | 0.462149 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
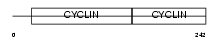
|
Domain Architecture |
|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CABYR_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.043663 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.030496 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.028761 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
GIDRP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.058286 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z5L2, O60598, Q5W0B4, Q5W0B5, Q86VT9, Q8N9H0 | Gene names | GIDRP88, C10orf28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 (Putative mitochondrial space protein 32.1). | |||||
|
U2AF2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.045220 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26368 | Gene names | U2AF2, U2AF65 | |||
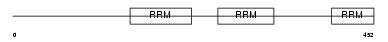
|
Domain Architecture |
|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.012273 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.015816 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
ATS17_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.006637 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE56 | Gene names | ADAMTS17 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-17 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 17) (ADAM-TS 17) (ADAM-TS17). | |||||
|
CCNG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.271258 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.284733 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.012953 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ARX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.003975 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
MKNK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.001463 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDB0, O08606, Q75PY0, Q9D893 | Gene names | Mknk2, Mnk2 | |||
|
Domain Architecture |
|
|||||
| Description | MAP kinase-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (MAP kinase signal-integrating kinase 2) (Mnk2). | |||||
|
MKNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.001182 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBH9, Q6GPI3, Q9HBH8, Q9UHR0, Q9Y2N6 | Gene names | MKNK2, MNK2 | |||
|
Domain Architecture |
|
|||||
| Description | MAP kinase-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (MAP kinase signal-integrating kinase 2) (Mnk2). | |||||
|
OSBP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.008250 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
PASK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.000457 (rank : 52) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.001923 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
MRP7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.004083 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4P9, Q3TCQ0, Q3V0D1, Q8R4S1 | Gene names | Abcc10, Mrp7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multidrug resistance-associated protein 7 (ATP-binding cassette sub- family C member 10). | |||||
|
SEL1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.023722 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.022547 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.003224 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
PSMD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.010782 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99460, Q53TI2, Q6GMU5, Q6P2P4, Q6PJM7, Q6PKG9, Q86VU1, Q8IV79 | Gene names | PSMD1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 1 (26S proteasome regulatory subunit RPN2) (26S proteasome regulatory subunit S1) (26S proteasome subunit p112). | |||||
|
PTN21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.002473 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
CCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.222842 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.223955 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
CCNI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.252339 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.232278 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-I. | |||||
|
CCNK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.070780 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.057510 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
UNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.195734 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CCNF_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_HUMAN
|
||||||
| NC score | 0.992573 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.731710 (rank : 3) | θ value | 1.6852e-22 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.730050 (rank : 4) | θ value | 2.20094e-22 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.727456 (rank : 5) | θ value | 1.86321e-21 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.720649 (rank : 6) | θ value | 1.74391e-19 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB1_HUMAN
|
||||||
| NC score | 0.702533 (rank : 7) | θ value | 1.80466e-16 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14635 | Gene names | CCNB1, CCNB | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB1_MOUSE
|
||||||
| NC score | 0.701730 (rank : 8) | θ value | 3.40345e-15 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24860 | Gene names | Ccnb1, Ccn-2, Ccnb1-rs13, Cycb, Cycb1 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B1. | |||||
|
CCNB2_MOUSE
|
||||||
| NC score | 0.693354 (rank : 9) | θ value | 1.99529e-15 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30276 | Gene names | Ccnb2, Cycb2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
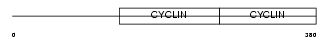
CCNB2_HUMAN
|
||||||
| NC score | 0.690962 (rank : 10) | θ value | 3.40345e-15 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95067 | Gene names | CCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B2. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.663048 (rank : 11) | θ value | 2.97466e-19 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.564845 (rank : 12) | θ value | 1.86753e-13 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCND1_MOUSE
|
||||||
| NC score | 0.543318 (rank : 13) | θ value | 2.61198e-07 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25322 | Gene names | Ccnd1, Cyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1. | |||||
|
CCNE2_MOUSE
|
||||||
| NC score | 0.542038 (rank : 14) | θ value | 2.21117e-06 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z238, Q9QWU5 | Gene names | Ccne2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCND1_HUMAN
|
||||||
| NC score | 0.541806 (rank : 15) | θ value | 8.97725e-08 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24385, Q6LEF0 | Gene names | CCND1, BCL1, PRAD1 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D1 (PRAD1 oncogene) (BCL-1 oncogene). | |||||
|
CCNE2_HUMAN
|
||||||
| NC score | 0.538753 (rank : 16) | θ value | 3.77169e-06 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O96020, O95439 | Gene names | CCNE2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E2. | |||||
|
CCNE1_HUMAN
|
||||||
| NC score | 0.523120 (rank : 17) | θ value | 0.000461057 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24864, Q14091, Q8NFG1, Q92501 | Gene names | CCNE1, CCNE | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCND2_HUMAN
|
||||||
| NC score | 0.520542 (rank : 18) | θ value | 1.69304e-06 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30279, Q13955 | Gene names | CCND2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCNE1_MOUSE
|
||||||
| NC score | 0.516113 (rank : 19) | θ value | 0.00134147 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61457 | Gene names | Ccne1, Ccne | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-E1. | |||||
|
CCND2_MOUSE
|
||||||
| NC score | 0.506121 (rank : 20) | θ value | 3.77169e-06 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30280 | Gene names | Ccnd2, Cyl-2 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D2. | |||||
|
CCND3_HUMAN
|
||||||
| NC score | 0.489197 (rank : 21) | θ value | 0.00390308 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P30281, Q5T8J0, Q96F49 | Gene names | CCND3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCND3_MOUSE
|
||||||
| NC score | 0.462149 (rank : 22) | θ value | 0.0113563 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30282, Q62391, Q99KP2 | Gene names | Ccnd3, Cyl-3 | |||
|
Domain Architecture |

|
|||||
| Description | G1/S-specific cyclin-D3. | |||||
|
CCNG1_MOUSE
|
||||||
| NC score | 0.284733 (rank : 23) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51945, O54779 | Gene names | Ccng1, Ccng | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNG1_HUMAN
|
||||||
| NC score | 0.271258 (rank : 24) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51959, Q15757, Q96L32 | Gene names | CCNG1, CCNG, CYCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G1 (Cyclin-G). | |||||
|
CCNI_HUMAN
|
||||||
| NC score | 0.252339 (rank : 25) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14094 | Gene names | CCNI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNI_MOUSE
|
||||||
| NC score | 0.232278 (rank : 26) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z2V9 | Gene names | Ccni | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-I. | |||||
|
CCNG2_MOUSE
|
||||||
| NC score | 0.223955 (rank : 27) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_HUMAN
|
||||||
| NC score | 0.222842 (rank : 28) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
UNG2_HUMAN
|
||||||
| NC score | 0.195734 (rank : 29) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.070780 (rank : 30) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
GIDRP_HUMAN
|
||||||
| NC score | 0.058286 (rank : 31) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z5L2, O60598, Q5W0B4, Q5W0B5, Q86VT9, Q8N9H0 | Gene names | GIDRP88, C10orf28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 (Putative mitochondrial space protein 32.1). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.057510 (rank : 32) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
U2AF2_HUMAN
|
||||||
| NC score | 0.045220 (rank : 33) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26368 | Gene names | U2AF2, U2AF65 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor U2AF 65 kDa subunit (U2 auxiliary factor 65 kDa subunit) (U2 snRNP auxiliary factor large subunit) (hU2AF(65)). | |||||
|
CABYR_MOUSE
|
||||||
| NC score | 0.043663 (rank : 34) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.030496 (rank : 35) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.028761 (rank : 36) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.023722 (rank : 37) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.022547 (rank : 38) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.015816 (rank : 39) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.012953 (rank : 40) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.012273 (rank : 41) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
PSMD1_HUMAN
|
||||||
| NC score | 0.010782 (rank : 42) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99460, Q53TI2, Q6GMU5, Q6P2P4, Q6PJM7, Q6PKG9, Q86VU1, Q8IV79 | Gene names | PSMD1 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 1 (26S proteasome regulatory subunit RPN2) (26S proteasome regulatory subunit S1) (26S proteasome subunit p112). | |||||
|
OSBP2_HUMAN
|
||||||
| NC score | 0.008250 (rank : 43) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q969R2, O60396, Q9BY96, Q9BZF0 | Gene names | OSBP2, KIAA1664, ORP4, OSBPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 2 (Oxysterol-binding protein-related protein 4) (OSBP-related protein 4) (ORP-4). | |||||
|
ATS17_HUMAN
|
||||||
| NC score | 0.006637 (rank : 44) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TE56 | Gene names | ADAMTS17 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-17 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 17) (ADAM-TS 17) (ADAM-TS17). | |||||
|
MRP7_MOUSE
|
||||||
| NC score | 0.004083 (rank : 45) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R4P9, Q3TCQ0, Q3V0D1, Q8R4S1 | Gene names | Abcc10, Mrp7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multidrug resistance-associated protein 7 (ATP-binding cassette sub- family C member 10). | |||||
|
ARX_HUMAN
|
||||||
| NC score | 0.003975 (rank : 46) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.003224 (rank : 47) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
PTN21_HUMAN
|
||||||
| NC score | 0.002473 (rank : 48) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16825 | Gene names | PTPN21, PTPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase D1). | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.001923 (rank : 49) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
MKNK2_MOUSE
|
||||||
| NC score | 0.001463 (rank : 50) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CDB0, O08606, Q75PY0, Q9D893 | Gene names | Mknk2, Mnk2 | |||
|
Domain Architecture |

|
|||||
| Description | MAP kinase-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (MAP kinase signal-integrating kinase 2) (Mnk2). | |||||
|
MKNK2_HUMAN
|
||||||
| NC score | 0.001182 (rank : 51) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBH9, Q6GPI3, Q9HBH8, Q9UHR0, Q9Y2N6 | Gene names | MKNK2, MNK2 | |||
|
Domain Architecture |

|
|||||
| Description | MAP kinase-interacting serine/threonine-protein kinase 2 (EC 2.7.11.1) (MAP kinase signal-integrating kinase 2) (Mnk2). | |||||
|
PASK_MOUSE
|
||||||
| NC score | 0.000457 (rank : 52) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||