Please be patient as the page loads
|
MYCL1_MOUSE
|
||||||
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MYCL1_MOUSE
|
||||||
| θ value | 1.41594e-162 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 4.12933e-154 (rank : 2) | NC score | 0.977155 (rank : 2) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | 1.2549e-118 (rank : 3) | NC score | 0.938542 (rank : 3) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYC_HUMAN
|
||||||
| θ value | 4.9032e-22 (rank : 4) | NC score | 0.749576 (rank : 5) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 5) | NC score | 0.750381 (rank : 4) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 6) | NC score | 0.742050 (rank : 7) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 7) | NC score | 0.743898 (rank : 6) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 8) | NC score | 0.702674 (rank : 8) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYCB_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 9) | NC score | 0.569292 (rank : 9) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MAX_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 10) | NC score | 0.546572 (rank : 10) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 11) | NC score | 0.543959 (rank : 11) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 12) | NC score | 0.327287 (rank : 12) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 13) | NC score | 0.202659 (rank : 34) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 14) | NC score | 0.199013 (rank : 36) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.315636 (rank : 13) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.187568 (rank : 43) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 17) | NC score | 0.188080 (rank : 42) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 18) | NC score | 0.271376 (rank : 22) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 19) | NC score | 0.281312 (rank : 20) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.231420 (rank : 28) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 21) | NC score | 0.040210 (rank : 126) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 22) | NC score | 0.216341 (rank : 32) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.238235 (rank : 25) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 24) | NC score | 0.295988 (rank : 15) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 25) | NC score | 0.298206 (rank : 14) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.287153 (rank : 19) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.165758 (rank : 44) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.049699 (rank : 121) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.030375 (rank : 136) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MLX_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.233032 (rank : 27) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.235145 (rank : 26) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | 0.125558 (rank : 32) | NC score | 0.196407 (rank : 37) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | 0.163984 (rank : 33) | NC score | 0.189271 (rank : 41) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.292473 (rank : 17) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
NGN3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.119447 (rank : 62) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.229297 (rank : 29) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.244589 (rank : 23) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
CBL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.031334 (rank : 134) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.066119 (rank : 100) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.066045 (rank : 101) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.227786 (rank : 30) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.293780 (rank : 16) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.291016 (rank : 18) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.044291 (rank : 123) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.274319 (rank : 21) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.239953 (rank : 24) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
TBCD2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.041652 (rank : 124) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.146887 (rank : 49) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.146879 (rank : 50) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.035191 (rank : 129) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CCNF_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.030496 (rank : 135) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
GTF2I_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.033424 (rank : 130) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
IPP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.048652 (rank : 122) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERT9 | Gene names | Ppp1r1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase inhibitor 1 (IPP-1) (I-1). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.010544 (rank : 158) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.019307 (rank : 144) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.115026 (rank : 64) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.111931 (rank : 65) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
KI13B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.014494 (rank : 150) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
NGN3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.104598 (rank : 66) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |
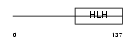
|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.192659 (rank : 39) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.193597 (rank : 38) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TWST1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 62) | NC score | 0.144210 (rank : 54) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 63) | NC score | 0.144213 (rank : 53) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.032945 (rank : 131) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
SCX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.123963 (rank : 61) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |
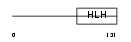
|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.012751 (rank : 154) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
TAF1C_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.032332 (rank : 132) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.131344 (rank : 57) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.130718 (rank : 58) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
HAND2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.149438 (rank : 46) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
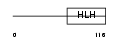
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.149438 (rank : 47) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
MITF_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.215315 (rank : 33) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
NGN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.097286 (rank : 69) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.098136 (rank : 68) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
PTF1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.145824 (rank : 51) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
PTF1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.142368 (rank : 56) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
ASCL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.155859 (rank : 45) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.031343 (rank : 133) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.217058 (rank : 31) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
NACA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.039425 (rank : 128) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13765, Q53A18, Q53G46 | Gene names | NACA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (NAC-alpha) (Alpha-NAC) (Hom s 2.02). | |||||
|
NACA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.039460 (rank : 127) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60817 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (Alpha-NAC) (Alpha-NAC/1.9.2). | |||||
|
CJ030_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.025998 (rank : 138) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N7W2, Q5SYY7, Q5SYY8, Q5SYY9, Q8N5T7 | Gene names | C10orf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf30. | |||||
|
KI67_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.040998 (rank : 125) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.028047 (rank : 137) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
VEGFB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.024818 (rank : 140) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49766, Q3UG04, Q5D0B1, Q64290 | Gene names | Vegfb, Vrf | |||
|
Domain Architecture |
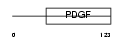
|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.149253 (rank : 48) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.025328 (rank : 139) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
MGRN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.020427 (rank : 143) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
NFIP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.024543 (rank : 141) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NV92, Q7Z2H3, Q7Z428, Q8TAR3, Q9ULQ5 | Gene names | NDFIP2, KIAA1165, N4WBP5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD4 family-interacting protein 2 (NEDD4 WW domain-binding protein 5A) (Putative MAPK-activating protein PM04/PM05/PM06/PM07) (Putative NF-kappa-B-activating protein 413). | |||||
|
NUMB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.016583 (rank : 148) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.009860 (rank : 159) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.005658 (rank : 164) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
ZFP95_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | -0.002501 (rank : 170) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |
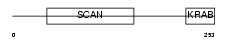
|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.000593 (rank : 169) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.144472 (rank : 52) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
ASCL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.119269 (rank : 63) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |
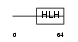
|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
ATS14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.004382 (rank : 168) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.092534 (rank : 71) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.092237 (rank : 72) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.091545 (rank : 73) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.092621 (rank : 70) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.006250 (rank : 163) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.009026 (rank : 161) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
GPX6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.009699 (rank : 160) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59796, Q4PJ17 | Gene names | GPX6 | |||
|
Domain Architecture |
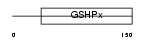
|
|||||
| Description | Glutathione peroxidase 6 precursor (EC 1.11.1.9). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.018163 (rank : 147) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.015940 (rank : 149) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.011663 (rank : 155) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
BACH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.018531 (rank : 145) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91V12, Q59HQ2 | Gene names | Acot7, Bach | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
GIPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.004591 (rank : 167) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48546, Q14401, Q16400, Q9UPI1 | Gene names | GIPR | |||
|
Domain Architecture |

|
|||||
| Description | Gastric inhibitory polypeptide receptor precursor (GIP-R) (Glucose- dependent insulinotropic polypeptide receptor). | |||||
|
HAND1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.124110 (rank : 60) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |
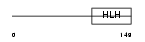
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.129195 (rank : 59) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.005497 (rank : 166) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.022328 (rank : 142) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.082784 (rank : 78) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.081913 (rank : 79) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.018213 (rank : 146) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.005522 (rank : 165) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.012920 (rank : 152) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
FRAT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.010628 (rank : 157) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |
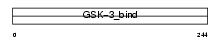
|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
I10R2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.010988 (rank : 156) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61190 | Gene names | Il10rb, Crfb4 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-10 receptor beta chain precursor (IL-10R-B) (IL-10R2) (Cytokine receptor family 2 member 4) (Cytokine receptor class-II member 4) (CRF2-4) (CDw210b antigen). | |||||
|
JHD2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.007522 (rank : 162) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
LIMA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.012752 (rank : 153) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.014403 (rank : 151) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
ASCL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.087672 (rank : 74) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |
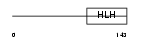
|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
ASCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.083637 (rank : 77) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
ATOH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.085671 (rank : 75) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ATOH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.084865 (rank : 76) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.050982 (rank : 118) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.050907 (rank : 119) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HEN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.076072 (rank : 85) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |
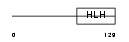
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.076827 (rank : 84) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |
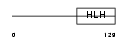
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.074056 (rank : 86) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |
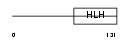
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.073694 (rank : 87) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |
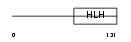
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.060244 (rank : 110) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.060790 (rank : 109) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.065074 (rank : 103) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.066365 (rank : 99) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
ID1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.053132 (rank : 117) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |
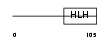
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
ID1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.050338 (rank : 120) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |
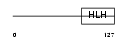
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
LYL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.068199 (rank : 95) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
LYL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.066499 (rank : 98) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.142503 (rank : 55) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.190568 (rank : 40) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MUSC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.064029 (rank : 106) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
MUSC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.065264 (rank : 102) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
MYF5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.056569 (rank : 114) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |
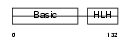
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.058256 (rank : 112) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |
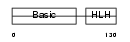
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.071310 (rank : 89) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
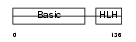
|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.068280 (rank : 94) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
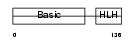
|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
MYOD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.054799 (rank : 116) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.055808 (rank : 115) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYOG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.064831 (rank : 104) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |
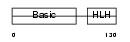
|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.064495 (rank : 105) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |
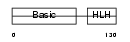
|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.056742 (rank : 113) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NDF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.059401 (rank : 111) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
NDF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.079663 (rank : 81) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.079498 (rank : 83) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NGN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.068178 (rank : 96) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |
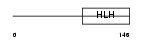
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
NGN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.069663 (rank : 92) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |
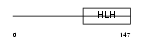
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.071211 (rank : 90) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.072036 (rank : 88) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.069162 (rank : 93) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
TAL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.067108 (rank : 97) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
TCF15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.079543 (rank : 82) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
TCF15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.081491 (rank : 80) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
TCF21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.063031 (rank : 107) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |
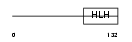
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.062545 (rank : 108) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |
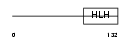
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.070082 (rank : 91) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.202529 (rank : 35) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.099159 (rank : 67) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.41594e-162 (rank : 1) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 123 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.977155 (rank : 2) | θ value | 4.12933e-154 (rank : 2) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.938542 (rank : 3) | θ value | 1.2549e-118 (rank : 3) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.750381 (rank : 4) | θ value | 7.0802e-21 (rank : 5) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.749576 (rank : 5) | θ value | 4.9032e-22 (rank : 4) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.743898 (rank : 6) | θ value | 5.07402e-19 (rank : 7) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.742050 (rank : 7) | θ value | 1.33526e-19 (rank : 6) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.702674 (rank : 8) | θ value | 3.52202e-12 (rank : 8) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.569292 (rank : 9) | θ value | 2.21117e-06 (rank : 9) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.546572 (rank : 10) | θ value | 4.92598e-06 (rank : 10) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.543959 (rank : 11) | θ value | 4.92598e-06 (rank : 11) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.327287 (rank : 12) | θ value | 0.00035302 (rank : 12) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.315636 (rank : 13) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.298206 (rank : 14) | θ value | 0.0252991 (rank : 25) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.295988 (rank : 15) | θ value | 0.0252991 (rank : 24) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.293780 (rank : 16) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.292473 (rank : 17) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.291016 (rank : 18) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.287153 (rank : 19) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.281312 (rank : 20) | θ value | 0.00228821 (rank : 19) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.274319 (rank : 21) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.271376 (rank : 22) | θ value | 0.00228821 (rank : 18) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.244589 (rank : 23) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.239953 (rank : 24) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.238235 (rank : 25) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.235145 (rank : 26) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.233032 (rank : 27) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.231420 (rank : 28) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.229297 (rank : 29) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.227786 (rank : 30) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.217058 (rank : 31) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.216341 (rank : 32) | θ value | 0.0193708 (rank : 22) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.215315 (rank : 33) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.202659 (rank : 34) | θ value | 0.000786445 (rank : 13) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.202529 (rank : 35) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.199013 (rank : 36) | θ value | 0.000786445 (rank : 14) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.196407 (rank : 37) | θ value | 0.125558 (rank : 32) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.193597 (rank : 38) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.192659 (rank : 39) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.190568 (rank : 40) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.189271 (rank : 41) | θ value | 0.163984 (rank : 33) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.188080 (rank : 42) | θ value | 0.00175202 (rank : 17) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.187568 (rank : 43) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.165758 (rank : 44) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
ASCL1_HUMAN
|
||||||
| NC score | 0.155859 (rank : 45) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.149438 (rank : 46) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.149438 (rank : 47) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.149253 (rank : 48) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.146887 (rank : 49) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.146879 (rank : 50) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
PTF1A_HUMAN
|
||||||
| NC score | 0.145824 (rank : 51) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.144472 (rank : 52) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
TWST1_MOUSE
|
||||||
| NC score | 0.144213 (rank : 53) | θ value | 0.62314 (rank : 63) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
TWST1_HUMAN
|
||||||
| NC score | 0.144210 (rank : 54) | θ value | 0.62314 (rank : 62) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.142503 (rank : 55) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
PTF1A_MOUSE
|
||||||
| NC score | 0.142368 (rank : 56) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.131344 (rank : 57) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.130718 (rank : 58) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.129195 (rank : 59) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
HAND1_HUMAN
|
||||||
| NC score | 0.124110 (rank : 60) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
SCX_MOUSE
|
||||||
| NC score | 0.123963 (rank : 61) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |

|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
NGN3_HUMAN
|
||||||
| NC score | 0.119447 (rank : 62) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
ASCL2_HUMAN
|
||||||
| NC score | 0.119269 (rank : 63) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.115026 (rank : 64) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.111931 (rank : 65) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
NGN3_MOUSE
|
||||||
| NC score | 0.104598 (rank : 66) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.099159 (rank : 67) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.098136 (rank : 68) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
NGN2_HUMAN
|
||||||
| NC score | 0.097286 (rank : 69) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.092621 (rank : 70) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.092534 (rank : 71) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.092237 (rank : 72) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.091545 (rank : 73) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
ASCL3_HUMAN
|
||||||
| NC score | 0.087672 (rank : 74) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
ATOH1_HUMAN
|
||||||
| NC score | 0.085671 (rank : 75) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ATOH1_MOUSE
|
||||||
| NC score | 0.084865 (rank : 76) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
ASCL3_MOUSE
|
||||||
| NC score | 0.083637 (rank : 77) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.082784 (rank : 78) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.081913 (rank : 79) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
TCF15_MOUSE
|
||||||
| NC score | 0.081491 (rank : 80) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
NDF2_HUMAN
|
||||||
| NC score | 0.079663 (rank : 81) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
TCF15_HUMAN
|
||||||
| NC score | 0.079543 (rank : 82) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
NDF2_MOUSE
|
||||||
| NC score | 0.079498 (rank : 83) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
HEN1_MOUSE
|
||||||
| NC score | 0.076827 (rank : 84) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_HUMAN
|
||||||
| NC score | 0.076072 (rank : 85) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN2_HUMAN
|
||||||
| NC score | 0.074056 (rank : 86) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| NC score | 0.073694 (rank : 87) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.072036 (rank : 88) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.071310 (rank : 89) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.071211 (rank : 90) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.070082 (rank : 91) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
NGN1_MOUSE
|
||||||
| NC score | 0.069663 (rank : 92) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
TAL2_HUMAN
|
||||||
| NC score | 0.069162 (rank : 93) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.068280 (rank : 94) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
LYL1_HUMAN
|
||||||
| NC score | 0.068199 (rank : 95) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
NGN1_HUMAN
|
||||||
| NC score | 0.068178 (rank : 96) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
TAL2_MOUSE
|
||||||
| NC score | 0.067108 (rank : 97) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
LYL1_MOUSE
|
||||||
| NC score | 0.066499 (rank : 98) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.066365 (rank : 99) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.066119 (rank : 100) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.066045 (rank : 101) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
MUSC_MOUSE
|
||||||
| NC score | 0.065264 (rank : 102) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.065074 (rank : 103) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
MYOG_HUMAN
|
||||||
| NC score | 0.064831 (rank : 104) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| NC score | 0.064495 (rank : 105) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
MUSC_HUMAN
|
||||||
| NC score | 0.064029 (rank : 106) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
TCF21_HUMAN
|
||||||
| NC score | 0.063031 (rank : 107) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| NC score | 0.062545 (rank : 108) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.060790 (rank : 109) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.060244 (rank : 110) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.059401 (rank : 111) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
MYF5_MOUSE
|
||||||
| NC score | 0.058256 (rank : 112) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.056742 (rank : 113) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
MYF5_HUMAN
|
||||||
| NC score | 0.056569 (rank : 114) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.055808 (rank : 115) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYOD1_HUMAN
|
||||||
| NC score | 0.054799 (rank : 116) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
ID1_HUMAN
|
||||||
| NC score | 0.053132 (rank : 117) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.050982 (rank : 118) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.050907 (rank : 119) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
ID1_MOUSE
|
||||||
| NC score | 0.050338 (rank : 120) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.049699 (rank : 121) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
IPP1_MOUSE
|
||||||
| NC score | 0.048652 (rank : 122) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERT9 | Gene names | Ppp1r1a | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase inhibitor 1 (IPP-1) (I-1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.044291 (rank : 123) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TBCD2_HUMAN
|
||||||
| NC score | 0.041652 (rank : 124) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYX2 | Gene names | TBC1D2, PARIS1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 2 (Prostate antigen recognized and identified by SEREX) (PARIS-1). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.040998 (rank : 125) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.040210 (rank : 126) | θ value | 0.0148317 (rank : 21) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NACA_MOUSE
|
||||||
| NC score | 0.039460 (rank : 127) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60817 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (Alpha-NAC) (Alpha-NAC/1.9.2). | |||||
|
NACA_HUMAN
|
||||||
| NC score | 0.039425 (rank : 128) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13765, Q53A18, Q53G46 | Gene names | NACA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha (NAC-alpha) (Alpha-NAC) (Hom s 2.02). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.035191 (rank : 129) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
GTF2I_HUMAN
|
||||||
| NC score | 0.033424 (rank : 130) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78347, O14743, O15359, O43546, O43588, O43589, Q9BSZ4 | Gene names | GTF2I, BAP135, WBSCR6 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I (GTFII-I) (TFII-I) (Bruton tyrosine kinase-associated protein 135) (BTK-associated protein 135) (BAP-135) (SRF-Phox1-interacting protein) (SPIN) (Williams-Beuren syndrome chromosome region 6 protein). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.032945 (rank : 131) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
TAF1C_HUMAN
|
||||||
| NC score | 0.032332 (rank : 132) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15572, Q59F67, Q8N6V3 | Gene names | TAF1C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor RNA polymerase I subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 110 kDa) (TAFI110). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.031343 (rank : 133) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CBL_MOUSE
|
||||||
| NC score | 0.031334 (rank : 134) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
CCNF_MOUSE
|
||||||
| NC score | 0.030496 (rank : 135) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.030375 (rank : 136) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.028047 (rank : 137) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
CJ030_HUMAN
|
||||||
| NC score | 0.025998 (rank : 138) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N7W2, Q5SYY7, Q5SYY8, Q5SYY9, Q8N5T7 | Gene names | C10orf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf30. | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.025328 (rank : 139) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
VEGFB_MOUSE
|
||||||
| NC score | 0.024818 (rank : 140) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49766, Q3UG04, Q5D0B1, Q64290 | Gene names | Vegfb, Vrf | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
NFIP2_HUMAN
|
||||||
| NC score | 0.024543 (rank : 141) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NV92, Q7Z2H3, Q7Z428, Q8TAR3, Q9ULQ5 | Gene names | NDFIP2, KIAA1165, N4WBP5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEDD4 family-interacting protein 2 (NEDD4 WW domain-binding protein 5A) (Putative MAPK-activating protein PM04/PM05/PM06/PM07) (Putative NF-kappa-B-activating protein 413). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.022328 (rank : 142) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
MGRN1_HUMAN
|
||||||
| NC score | 0.020427 (rank : 143) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.019307 (rank : 144) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
BACH_MOUSE
|
||||||
| NC score | 0.018531 (rank : 145) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91V12, Q59HQ2 | Gene names | Acot7, Bach | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.018213 (rank : 146) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.018163 (rank : 147) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NUMB_MOUSE
|
||||||
| NC score | 0.016583 (rank : 148) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.015940 (rank : 149) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
KI13B_HUMAN
|
||||||
| NC score | 0.014494 (rank : 150) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NQT8, O75134, Q9BYJ6 | Gene names | KIF13B, GAKIN, KIAA0639 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13B (Kinesin-like protein GAKIN). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.014403 (rank : 151) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.012920 (rank : 152) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
LIMA1_MOUSE
|
||||||
| NC score | 0.012752 (rank : 153) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.012751 (rank : 154) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.011663 (rank : 155) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
I10R2_MOUSE
|
||||||
| NC score | 0.010988 (rank : 156) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61190 | Gene names | Il10rb, Crfb4 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-10 receptor beta chain precursor (IL-10R-B) (IL-10R2) (Cytokine receptor family 2 member 4) (Cytokine receptor class-II member 4) (CRF2-4) (CDw210b antigen). | |||||
|
FRAT1_MOUSE
|
||||||
| NC score | 0.010628 (rank : 157) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.010544 (rank : 158) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.009860 (rank : 159) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
GPX6_HUMAN
|
||||||
| NC score | 0.009699 (rank : 160) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59796, Q4PJ17 | Gene names | GPX6 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione peroxidase 6 precursor (EC 1.11.1.9). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.009026 (rank : 161) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.007522 (rank : 162) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.006250 (rank : 163) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.005658 (rank : 164) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.005522 (rank : 165) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.005497 (rank : 166) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
GIPR_HUMAN
|
||||||
| NC score | 0.004591 (rank : 167) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48546, Q14401, Q16400, Q9UPI1 | Gene names | GIPR | |||
|
Domain Architecture |

|
|||||
| Description | Gastric inhibitory polypeptide receptor precursor (GIP-R) (Glucose- dependent insulinotropic polypeptide receptor). | |||||
|
ATS14_HUMAN
|
||||||
| NC score | 0.004382 (rank : 168) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.000593 (rank : 169) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ZFP95_HUMAN
|
||||||
| NC score | -0.002501 (rank : 170) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 123 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||