Please be patient as the page loads
|
MAD1_HUMAN
|
||||||
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MAD1_HUMAN
|
||||||
| θ value | 1.59853e-89 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 1.18825e-76 (rank : 2) | NC score | 0.984508 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 1.19932e-44 (rank : 3) | NC score | 0.924945 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 5.03882e-43 (rank : 4) | NC score | 0.933012 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 6.37413e-38 (rank : 5) | NC score | 0.905169 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 3.4976e-36 (rank : 6) | NC score | 0.919669 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 1.02001e-27 (rank : 7) | NC score | 0.890053 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 8) | NC score | 0.890323 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 9) | NC score | 0.477215 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 10) | NC score | 0.459249 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
NGN3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.085685 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
MAX_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.240313 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.234987 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.179008 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.192116 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
NGN3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.085290 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |
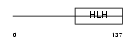
|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.216170 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.164345 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.185123 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.140469 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.140389 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.236783 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.235597 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
CING_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.024445 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.112614 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.021229 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.157124 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
USF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.226917 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.227185 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.012944 (rank : 80) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.030381 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.027838 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.022218 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
CING_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.028656 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.148430 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
NGN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.052005 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.050889 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.019493 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.053062 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.073509 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
CL011_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.038032 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NVM9, Q86WE2, Q96HM2, Q9BTX2, Q9NTB6, Q9NVM5 | Gene names | C12orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf11 (Sarcoma antigen NY-SAR-95). | |||||
|
HES4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.112237 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.075443 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
CL011_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.037023 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZV7, Q99JT8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf11 homolog. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.017790 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.072107 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CF152_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.018175 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.045241 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
ZN746_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.000902 (rank : 85) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U133 | Gene names | Znf746, Zfp746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.056980 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.048879 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.013305 (rank : 79) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.019274 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
NMDE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.006157 (rank : 83) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
NRBF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.040028 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96F24, Q86UR2, Q96NP6, Q9H0S9, Q9H2I2 | Gene names | NRBF2, COPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2) (Comodulator of PPAR and RXR). | |||||
|
CENPQ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.022779 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L2Z9, Q6IN61, Q9NVS5 | Gene names | CENPQ, C6orf139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.051334 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
HES2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.076103 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.112628 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.010148 (rank : 81) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.162817 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
ZN746_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.000468 (rank : 86) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NUN9 | Gene names | ZNF746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
CF060_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.015652 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
K1802_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.003265 (rank : 84) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.023218 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
RABE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.030114 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.017429 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TM9S4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.008210 (rank : 82) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92544, Q9NUA3 | Gene names | TM9SF4, KIAA0255 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane 9 superfamily protein member 4. | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052424 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
HES3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.050339 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.051111 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.052854 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.054133 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.056671 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.056863 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.140554 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.142503 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.164398 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.143353 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.148387 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.201162 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.205753 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.092572 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.066518 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.114209 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.114541 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.59853e-89 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.984508 (rank : 2) | θ value | 1.18825e-76 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.933012 (rank : 3) | θ value | 5.03882e-43 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.924945 (rank : 4) | θ value | 1.19932e-44 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.919669 (rank : 5) | θ value | 3.4976e-36 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.905169 (rank : 6) | θ value | 6.37413e-38 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.890323 (rank : 7) | θ value | 3.28125e-26 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.890053 (rank : 8) | θ value | 1.02001e-27 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.477215 (rank : 9) | θ value | 4.76016e-09 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.459249 (rank : 10) | θ value | 1.80886e-08 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.240313 (rank : 11) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.236783 (rank : 12) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.235597 (rank : 13) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.234987 (rank : 14) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.227185 (rank : 15) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.226917 (rank : 16) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.216170 (rank : 17) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.205753 (rank : 18) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.201162 (rank : 19) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.192116 (rank : 20) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.185123 (rank : 21) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.179008 (rank : 22) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.164398 (rank : 23) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.164345 (rank : 24) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.162817 (rank : 25) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.157124 (rank : 26) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.148430 (rank : 27) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.148387 (rank : 28) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.143353 (rank : 29) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.142503 (rank : 30) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.140554 (rank : 31) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.140469 (rank : 32) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.140389 (rank : 33) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.114541 (rank : 34) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.114209 (rank : 35) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.112628 (rank : 36) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.112614 (rank : 37) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.112237 (rank : 38) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.092572 (rank : 39) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
NGN3_HUMAN
|
||||||
| NC score | 0.085685 (rank : 40) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
NGN3_MOUSE
|
||||||
| NC score | 0.085290 (rank : 41) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.076103 (rank : 42) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.075443 (rank : 43) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.073509 (rank : 44) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.072107 (rank : 45) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.066518 (rank : 46) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.056980 (rank : 47) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.056863 (rank : 48) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.056671 (rank : 49) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.054133 (rank : 50) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.053062 (rank : 51) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.052854 (rank : 52) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.052424 (rank : 53) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
NGN2_HUMAN
|
||||||
| NC score | 0.052005 (rank : 54) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.051334 (rank : 55) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.051111 (rank : 56) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.050889 (rank : 57) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.050339 (rank : 58) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.048879 (rank : 59) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.045241 (rank : 60) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
NRBF2_HUMAN
|
||||||
| NC score | 0.040028 (rank : 61) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96F24, Q86UR2, Q96NP6, Q9H0S9, Q9H2I2 | Gene names | NRBF2, COPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2) (Comodulator of PPAR and RXR). | |||||
|
CL011_HUMAN
|
||||||
| NC score | 0.038032 (rank : 62) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NVM9, Q86WE2, Q96HM2, Q9BTX2, Q9NTB6, Q9NVM5 | Gene names | C12orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf11 (Sarcoma antigen NY-SAR-95). | |||||
|
CL011_MOUSE
|
||||||
| NC score | 0.037023 (rank : 63) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZV7, Q99JT8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf11 homolog. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.030381 (rank : 64) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
RABE2_MOUSE
|
||||||
| NC score | 0.030114 (rank : 65) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.028656 (rank : 66) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.027838 (rank : 67) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.024445 (rank : 68) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.023218 (rank : 69) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CENPQ_HUMAN
|
||||||
| NC score | 0.022779 (rank : 70) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L2Z9, Q6IN61, Q9NVS5 | Gene names | CENPQ, C6orf139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.022218 (rank : 71) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.021229 (rank : 72) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.019493 (rank : 73) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.019274 (rank : 74) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.018175 (rank : 75) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.017790 (rank : 76) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.017429 (rank : 77) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.015652 (rank : 78) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.013305 (rank : 79) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.012944 (rank : 80) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.010148 (rank : 81) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
TM9S4_HUMAN
|
||||||
| NC score | 0.008210 (rank : 82) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92544, Q9NUA3 | Gene names | TM9SF4, KIAA0255 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane 9 superfamily protein member 4. | |||||
|
NMDE1_HUMAN
|
||||||
| NC score | 0.006157 (rank : 83) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12879, O00669 | Gene names | GRIN2A | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 1 precursor (N-methyl D- aspartate receptor subtype 2A) (NR2A) (NMDAR2A) (hNR2A). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.003265 (rank : 84) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ZN746_MOUSE
|
||||||
| NC score | 0.000902 (rank : 85) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U133 | Gene names | Znf746, Zfp746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
ZN746_HUMAN
|
||||||
| NC score | 0.000468 (rank : 86) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NUN9 | Gene names | ZNF746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||