Please be patient as the page loads
|
HES1_MOUSE
|
||||||
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HES1_HUMAN
|
||||||
| θ value | 3.77839e-115 (rank : 1) | NC score | 0.995001 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 1.81628e-109 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES4_HUMAN
|
||||||
| θ value | 1.56274e-52 (rank : 3) | NC score | 0.977661 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HES2_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 4) | NC score | 0.884414 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 5) | NC score | 0.892278 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 6) | NC score | 0.789845 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 7) | NC score | 0.787638 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 8) | NC score | 0.771523 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HES6_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 9) | NC score | 0.799255 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 10) | NC score | 0.774446 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HES6_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 11) | NC score | 0.789103 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
HES5_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 12) | NC score | 0.821829 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HES7_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 13) | NC score | 0.701064 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 14) | NC score | 0.703848 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 15) | NC score | 0.585492 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 16) | NC score | 0.580127 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 17) | NC score | 0.581614 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 18) | NC score | 0.563815 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
HES3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 19) | NC score | 0.687323 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 20) | NC score | 0.234860 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.233812 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 22) | NC score | 0.228803 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.162689 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.187460 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.139851 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.142142 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.024478 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.121685 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYOD1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.114725 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.115489 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.149368 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYOG_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.112735 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.111820 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |
|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.032320 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.198169 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.198292 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.140389 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.024687 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.195447 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.196051 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.019618 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.070355 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.070342 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.183361 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.005342 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.024537 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MYF6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.100453 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
|
|||||
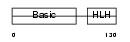
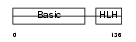
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.099925 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
|
|||||
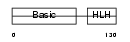
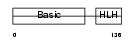
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.171608 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.185868 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.016505 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.036211 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.016642 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.009084 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.023657 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.154735 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.033449 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.120007 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.118710 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.016511 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
P2RX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.013615 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBL9, Q6V9R6, Q9NR37, Q9NR38, Q9UHD5, Q9UHD6, Q9UHD7, Q9Y637, Q9Y638 | Gene names | P2RX2, P2X2 | |||
|
Domain Architecture |
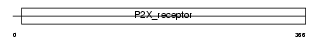
|
|||||
| Description | P2X purinoceptor 2 (ATP receptor) (P2X2) (Purinergic receptor). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.005626 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
TMEPA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.031277 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7R2, Q9EQH9 | Gene names | Tmepai, N4wbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane prostate androgen-induced protein (NEDD4 WW domain- binding protein 4). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.013555 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.010833 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.013759 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.019301 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.010324 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
G3ST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.011583 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RP7, Q8N3P7, Q8WZ17, Q96E33, Q9HA78 | Gene names | GAL3ST4 | |||
|
Domain Architecture |
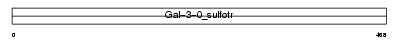
|
|||||
| Description | Galactose-3-O-sulfotransferase 4 (EC 2.8.2.-) (Galbeta1-3GalNAc 3'- sulfotransferase) (Beta-galactose-3-O-sulfotransferase 4). | |||||
|
IRK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.003961 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48549, Q8TBI0 | Gene names | KCNJ3, GIRK1 | |||
|
Domain Architecture |
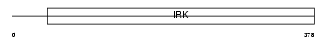
|
|||||
| Description | G protein-activated inward rectifier potassium channel 1 (GIRK1) (Potassium channel, inwardly rectifying subfamily J member 3) (Inward rectifier K(+) channel Kir3.1). | |||||
|
MYF5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.073288 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |
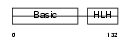
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.072509 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |
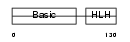
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.003133 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.002297 (rank : 90) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.028742 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.006089 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | -0.002359 (rank : 91) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZN692_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | -0.002370 (rank : 92) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BU19, Q5SRA5, Q5SRA6, Q9HBC9, Q9NW93, Q9NWY6, Q9UF97 | Gene names | ZNF692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.073803 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.075318 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.076331 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.076143 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.062299 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.060693 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.093715 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.081705 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.072146 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.073005 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.126264 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.129535 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.053801 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.057071 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.81628e-109 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.995001 (rank : 2) | θ value | 3.77839e-115 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.977661 (rank : 3) | θ value | 1.56274e-52 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.892278 (rank : 4) | θ value | 1.86321e-21 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_HUMAN
|
||||||
| NC score | 0.884414 (rank : 5) | θ value | 1.86321e-21 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.821829 (rank : 6) | θ value | 1.74796e-11 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.799255 (rank : 7) | θ value | 1.80466e-16 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.789845 (rank : 8) | θ value | 5.07402e-19 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HES6_MOUSE
|
||||||
| NC score | 0.789103 (rank : 9) | θ value | 6.85773e-16 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.787638 (rank : 10) | θ value | 5.60996e-18 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.774446 (rank : 11) | θ value | 4.02038e-16 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.771523 (rank : 12) | θ value | 6.20254e-17 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.703848 (rank : 13) | θ value | 9.59137e-10 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.701064 (rank : 14) | θ value | 2.52405e-10 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.687323 (rank : 15) | θ value | 1.17247e-07 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.585492 (rank : 16) | θ value | 4.76016e-09 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.581614 (rank : 17) | θ value | 1.80886e-08 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.580127 (rank : 18) | θ value | 1.06045e-08 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.563815 (rank : 19) | θ value | 5.26297e-08 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.234860 (rank : 20) | θ value | 0.00298849 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.233812 (rank : 21) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.228803 (rank : 22) | θ value | 0.0113563 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.198292 (rank : 23) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.198169 (rank : 24) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.196051 (rank : 25) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.195447 (rank : 26) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.187460 (rank : 27) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.185868 (rank : 28) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.183361 (rank : 29) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.171608 (rank : 30) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.162689 (rank : 31) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.154735 (rank : 32) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.149368 (rank : 33) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.142142 (rank : 34) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.140389 (rank : 35) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.139851 (rank : 36) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.129535 (rank : 37) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.126264 (rank : 38) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.121685 (rank : 39) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.120007 (rank : 40) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.118710 (rank : 41) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.115489 (rank : 42) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYOD1_HUMAN
|
||||||
| NC score | 0.114725 (rank : 43) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYOG_HUMAN
|
||||||
| NC score | 0.112735 (rank : 44) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| NC score | 0.111820 (rank : 45) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.100453 (rank : 46) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.099925 (rank : 47) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.093715 (rank : 48) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.081705 (rank : 49) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.076331 (rank : 50) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.076143 (rank : 51) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.075318 (rank : 52) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.073803 (rank : 53) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
MYF5_HUMAN
|
||||||
| NC score | 0.073288 (rank : 54) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.073005 (rank : 55) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MYF5_MOUSE
|
||||||
| NC score | 0.072509 (rank : 56) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.072146 (rank : 57) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.070355 (rank : 58) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.070342 (rank : 59) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.062299 (rank : 60) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.060693 (rank : 61) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.057071 (rank : 62) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.053801 (rank : 63) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.036211 (rank : 64) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.033449 (rank : 65) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.032320 (rank : 66) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
TMEPA_MOUSE
|
||||||
| NC score | 0.031277 (rank : 67) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7R2, Q9EQH9 | Gene names | Tmepai, N4wbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane prostate androgen-induced protein (NEDD4 WW domain- binding protein 4). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.028742 (rank : 68) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.024687 (rank : 69) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.024537 (rank : 70) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.024478 (rank : 71) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.023657 (rank : 72) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.019618 (rank : 73) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.019301 (rank : 74) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.016642 (rank : 75) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.016511 (rank : 76) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.016505 (rank : 77) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.013759 (rank : 78) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
P2RX2_HUMAN
|
||||||
| NC score | 0.013615 (rank : 79) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBL9, Q6V9R6, Q9NR37, Q9NR38, Q9UHD5, Q9UHD6, Q9UHD7, Q9Y637, Q9Y638 | Gene names | P2RX2, P2X2 | |||
|
Domain Architecture |
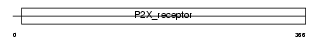
|
|||||
| Description | P2X purinoceptor 2 (ATP receptor) (P2X2) (Purinergic receptor). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.013555 (rank : 80) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
G3ST4_HUMAN
|
||||||
| NC score | 0.011583 (rank : 81) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RP7, Q8N3P7, Q8WZ17, Q96E33, Q9HA78 | Gene names | GAL3ST4 | |||
|
Domain Architecture |
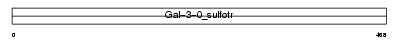
|
|||||
| Description | Galactose-3-O-sulfotransferase 4 (EC 2.8.2.-) (Galbeta1-3GalNAc 3'- sulfotransferase) (Beta-galactose-3-O-sulfotransferase 4). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.010833 (rank : 82) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.010324 (rank : 83) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.009084 (rank : 84) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.006089 (rank : 85) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.005626 (rank : 86) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.005342 (rank : 87) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
IRK3_HUMAN
|
||||||
| NC score | 0.003961 (rank : 88) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48549, Q8TBI0 | Gene names | KCNJ3, GIRK1 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 1 (GIRK1) (Potassium channel, inwardly rectifying subfamily J member 3) (Inward rectifier K(+) channel Kir3.1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.003133 (rank : 89) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.002297 (rank : 90) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | -0.002359 (rank : 91) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZN692_HUMAN
|
||||||
| NC score | -0.002370 (rank : 92) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BU19, Q5SRA5, Q5SRA6, Q9HBC9, Q9NW93, Q9NWY6, Q9UF97 | Gene names | ZNF692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||