Please be patient as the page loads
|
CLOCK_HUMAN
|
||||||
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CLOCK_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993703 (rank : 2) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.968979 (rank : 4) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.970057 (rank : 3) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 2.58786e-39 (rank : 5) | NC score | 0.856452 (rank : 5) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 3.73686e-38 (rank : 6) | NC score | 0.852751 (rank : 6) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 1.37539e-32 (rank : 7) | NC score | 0.811382 (rank : 7) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.28732e-30 (rank : 8) | NC score | 0.795021 (rank : 14) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 9) | NC score | 0.807441 (rank : 9) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 6.38894e-30 (rank : 10) | NC score | 0.809938 (rank : 8) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 11) | NC score | 0.803915 (rank : 10) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 1.57365e-28 (rank : 12) | NC score | 0.802272 (rank : 11) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 13) | NC score | 0.795797 (rank : 12) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 14) | NC score | 0.763848 (rank : 15) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 15) | NC score | 0.795088 (rank : 13) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 16) | NC score | 0.763832 (rank : 16) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 17) | NC score | 0.760858 (rank : 17) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 18) | NC score | 0.759126 (rank : 18) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 19) | NC score | 0.753697 (rank : 19) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 20) | NC score | 0.745754 (rank : 20) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 21) | NC score | 0.723335 (rank : 21) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 22) | NC score | 0.600776 (rank : 28) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 23) | NC score | 0.600434 (rank : 29) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 24) | NC score | 0.719957 (rank : 22) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 25) | NC score | 0.650059 (rank : 26) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 26) | NC score | 0.652972 (rank : 25) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 27) | NC score | 0.602801 (rank : 27) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 28) | NC score | 0.706412 (rank : 23) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 29) | NC score | 0.702971 (rank : 24) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 30) | NC score | 0.562598 (rank : 30) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 31) | NC score | 0.381942 (rank : 38) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 32) | NC score | 0.425765 (rank : 33) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 33) | NC score | 0.387481 (rank : 37) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 34) | NC score | 0.400939 (rank : 34) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 35) | NC score | 0.392100 (rank : 35) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 36) | NC score | 0.388744 (rank : 36) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 37) | NC score | 0.533149 (rank : 31) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 38) | NC score | 0.517707 (rank : 32) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 39) | NC score | 0.231805 (rank : 39) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 40) | NC score | 0.223297 (rank : 41) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 41) | NC score | 0.224181 (rank : 40) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 42) | NC score | 0.152587 (rank : 51) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 43) | NC score | 0.162829 (rank : 49) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MLX_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 44) | NC score | 0.192155 (rank : 45) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.145088 (rank : 52) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
MLX_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 46) | NC score | 0.192989 (rank : 44) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 47) | NC score | 0.206389 (rank : 43) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 48) | NC score | 0.206747 (rank : 42) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 49) | NC score | 0.056043 (rank : 76) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
K0240_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 50) | NC score | 0.041949 (rank : 82) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 51) | NC score | 0.036729 (rank : 87) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ZN185_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 52) | NC score | 0.065451 (rank : 71) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 53) | NC score | 0.045096 (rank : 80) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 54) | NC score | 0.061895 (rank : 74) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
EMIL3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.044098 (rank : 81) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NT22, Q76KT4 | Gene names | EMILIN3, C20orf130, EMILIN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMILIN-3 precursor (EMILIN-5) (Elastin microfibril interface-located protein 5) (Elastin microfibril interfacer 5). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.027901 (rank : 93) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 57) | NC score | 0.017087 (rank : 111) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.279714 (rank : 58) | NC score | 0.027218 (rank : 94) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CCD91_MOUSE
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.032237 (rank : 89) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.038126 (rank : 83) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.049560 (rank : 78) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.161967 (rank : 50) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.167005 (rank : 46) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 64) | NC score | 0.016015 (rank : 115) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 65) | NC score | 0.029212 (rank : 91) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 66) | NC score | 0.032466 (rank : 88) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 67) | NC score | 0.037906 (rank : 84) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.120811 (rank : 59) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.118888 (rank : 60) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
HES5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.121847 (rank : 58) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 71) | NC score | 0.166919 (rank : 47) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 72) | NC score | 0.164215 (rank : 48) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
EMIL3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.036907 (rank : 86) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59900 | Gene names | Emilin3, Emilin5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMILIN-3 precursor (EMILIN-5) (Elastin microfibril interface located protein 5) (Elastin microfibril interfacer 5). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.030664 (rank : 90) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.112250 (rank : 61) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
ZN148_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | -0.002623 (rank : 136) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||
|
HES3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.089257 (rank : 66) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.021359 (rank : 101) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.021046 (rank : 102) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.112203 (rank : 62) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.110722 (rank : 63) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
CENPQ_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.028510 (rank : 92) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7L2Z9, Q6IN61, Q9NVS5 | Gene names | CENPQ, C6orf139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.019889 (rank : 106) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.022310 (rank : 99) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.022123 (rank : 100) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYOC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.019815 (rank : 107) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70624, O70289 | Gene names | Myoc, Tigr | |||
|
Domain Architecture |
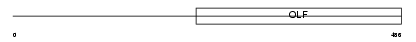
|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
NFKB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.010701 (rank : 123) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.025962 (rank : 95) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.020094 (rank : 105) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.003914 (rank : 133) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
PARP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.016903 (rank : 113) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
SH24A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.023413 (rank : 96) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.016976 (rank : 112) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.013021 (rank : 118) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.022624 (rank : 97) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MAX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.098276 (rank : 64) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.086422 (rank : 67) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.091545 (rank : 65) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.019559 (rank : 108) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.130633 (rank : 55) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.001191 (rank : 135) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.037133 (rank : 85) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
K0423_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.020160 (rank : 104) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.015921 (rank : 116) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
NARGL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.006982 (rank : 130) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
NFKB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.010125 (rank : 127) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.012963 (rank : 119) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
SH24A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.019438 (rank : 109) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.002813 (rank : 134) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.011248 (rank : 122) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DCP1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.020740 (rank : 103) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZD4, Q86XH9, Q96BP8, Q96MZ8 | Gene names | DCP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1B (EC 3.-.-.-). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.012331 (rank : 121) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.018930 (rank : 110) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.010613 (rank : 124) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.016596 (rank : 114) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.048477 (rank : 79) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.010055 (rank : 128) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BMF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.015746 (rank : 117) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96LC9, Q2M396, Q6NT30, Q6NT56, Q6P9F6, Q7Z7D4, Q7Z7D5, Q9H7K7 | Gene names | BMF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-modifying factor. | |||||
|
CLC4F_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.010242 (rank : 126) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N1N0 | Gene names | CLEC4F, CLECSF13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13). | |||||
|
FRS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.005342 (rank : 131) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
HES7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.083351 (rank : 68) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.079596 (rank : 69) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.012522 (rank : 120) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.010538 (rank : 125) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.022361 (rank : 98) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TOM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.004661 (rank : 132) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
UACA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.008937 (rank : 129) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
HES1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.061960 (rank : 73) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.062299 (rank : 72) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.068688 (rank : 70) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.059750 (rank : 75) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.052152 (rank : 77) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.136779 (rank : 54) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.136938 (rank : 53) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.130622 (rank : 56) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.124371 (rank : 57) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.993703 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.970057 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.968979 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.856452 (rank : 5) | θ value | 2.58786e-39 (rank : 5) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.852751 (rank : 6) | θ value | 3.73686e-38 (rank : 6) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.811382 (rank : 7) | θ value | 1.37539e-32 (rank : 7) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.809938 (rank : 8) | θ value | 6.38894e-30 (rank : 10) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.807441 (rank : 9) | θ value | 4.89182e-30 (rank : 9) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.803915 (rank : 10) | θ value | 8.3442e-30 (rank : 11) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.802272 (rank : 11) | θ value | 1.57365e-28 (rank : 12) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.795797 (rank : 12) | θ value | 1.16704e-23 (rank : 13) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.795088 (rank : 13) | θ value | 1.99067e-23 (rank : 15) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.795021 (rank : 14) | θ value | 1.28732e-30 (rank : 8) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.763848 (rank : 15) | θ value | 1.16704e-23 (rank : 14) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.763832 (rank : 16) | θ value | 3.39556e-23 (rank : 16) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.760858 (rank : 17) | θ value | 1.6852e-22 (rank : 17) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.759126 (rank : 18) | θ value | 2.87452e-22 (rank : 18) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.753697 (rank : 19) | θ value | 1.5773e-20 (rank : 19) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.745754 (rank : 20) | θ value | 2.06002e-20 (rank : 20) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.723335 (rank : 21) | θ value | 3.28887e-18 (rank : 21) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.719957 (rank : 22) | θ value | 7.32683e-18 (rank : 24) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.706412 (rank : 23) | θ value | 6.41864e-14 (rank : 28) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.702971 (rank : 24) | θ value | 1.86753e-13 (rank : 29) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.652972 (rank : 25) | θ value | 3.07829e-16 (rank : 26) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.650059 (rank : 26) | θ value | 3.07829e-16 (rank : 25) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.602801 (rank : 27) | θ value | 1.68911e-14 (rank : 27) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.600776 (rank : 28) | θ value | 7.32683e-18 (rank : 22) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.600434 (rank : 29) | θ value | 7.32683e-18 (rank : 23) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.562598 (rank : 30) | θ value | 6.64225e-11 (rank : 30) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.533149 (rank : 31) | θ value | 6.43352e-06 (rank : 37) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.517707 (rank : 32) | θ value | 6.43352e-06 (rank : 38) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.425765 (rank : 33) | θ value | 3.08544e-08 (rank : 32) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.400939 (rank : 34) | θ value | 3.41135e-07 (rank : 34) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.392100 (rank : 35) | θ value | 2.21117e-06 (rank : 35) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.388744 (rank : 36) | θ value | 2.21117e-06 (rank : 36) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.387481 (rank : 37) | θ value | 6.87365e-08 (rank : 33) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.381942 (rank : 38) | θ value | 2.36244e-08 (rank : 31) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.231805 (rank : 39) | θ value | 0.000121331 (rank : 39) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.224181 (rank : 40) | θ value | 0.00102713 (rank : 41) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.223297 (rank : 41) | θ value | 0.000786445 (rank : 40) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.206747 (rank : 42) | θ value | 0.00665767 (rank : 48) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.206389 (rank : 43) | θ value | 0.00665767 (rank : 47) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.192989 (rank : 44) | θ value | 0.00665767 (rank : 46) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.192155 (rank : 45) | θ value | 0.00390308 (rank : 44) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.167005 (rank : 46) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.166919 (rank : 47) | θ value | 0.62314 (rank : 71) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.164215 (rank : 48) | θ value | 0.62314 (rank : 72) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.162829 (rank : 49) | θ value | 0.00298849 (rank : 43) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.161967 (rank : 50) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.152587 (rank : 51) | θ value | 0.00228821 (rank : 42) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.145088 (rank : 52) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.136938 (rank : 53) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.136779 (rank : 54) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.130633 (rank : 55) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.130622 (rank : 56) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.124371 (rank : 57) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.121847 (rank : 58) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.120811 (rank : 59) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.118888 (rank : 60) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.112250 (rank : 61) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.112203 (rank : 62) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.110722 (rank : 63) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.098276 (rank : 64) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.091545 (rank : 65) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.089257 (rank : 66) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.086422 (rank : 67) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.083351 (rank : 68) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.079596 (rank : 69) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.068688 (rank : 70) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
ZN185_HUMAN
|
||||||
| NC score | 0.065451 (rank : 71) | θ value | 0.0563607 (rank : 52) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.062299 (rank : 72) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.061960 (rank : 73) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.061895 (rank : 74) | θ value | 0.0961366 (rank : 54) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.059750 (rank : 75) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.056043 (rank : 76) | θ value | 0.0330416 (rank : 49) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.052152 (rank : 77) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.049560 (rank : 78) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.048477 (rank : 79) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.045096 (rank : 80) | θ value | 0.0736092 (rank : 53) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
EMIL3_HUMAN
|
||||||
| NC score | 0.044098 (rank : 81) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NT22, Q76KT4 | Gene names | EMILIN3, C20orf130, EMILIN5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMILIN-3 precursor (EMILIN-5) (Elastin microfibril interface-located protein 5) (Elastin microfibril interfacer 5). | |||||
|
K0240_HUMAN
|
||||||
| NC score | 0.041949 (rank : 82) | θ value | 0.0431538 (rank : 50) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6AI39, Q5TFZ3, Q92514 | Gene names | KIAA0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.038126 (rank : 83) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.037906 (rank : 84) | θ value | 0.47712 (rank : 67) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.037133 (rank : 85) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
EMIL3_MOUSE
|
||||||
| NC score | 0.036907 (rank : 86) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P59900 | Gene names | Emilin3, Emilin5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EMILIN-3 precursor (EMILIN-5) (Elastin microfibril interface located protein 5) (Elastin microfibril interfacer 5). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.036729 (rank : 87) | θ value | 0.0563607 (rank : 51) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.032466 (rank : 88) | θ value | 0.47712 (rank : 66) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.032237 (rank : 89) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.030664 (rank : 90) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.029212 (rank : 91) | θ value | 0.47712 (rank : 65) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
CENPQ_HUMAN
|
||||||
| NC score | 0.028510 (rank : 92) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7L2Z9, Q6IN61, Q9NVS5 | Gene names | CENPQ, C6orf139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.027901 (rank : 93) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.027218 (rank : 94) | θ value | 0.279714 (rank : 58) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.025962 (rank : 95) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
SH24A_MOUSE
|
||||||
| NC score | 0.023413 (rank : 96) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.022624 (rank : 97) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.022361 (rank : 98) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.022310 (rank : 99) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.022123 (rank : 100) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.021359 (rank : 101) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.021046 (rank : 102) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
DCP1B_HUMAN
|
||||||
| NC score | 0.020740 (rank : 103) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZD4, Q86XH9, Q96BP8, Q96MZ8 | Gene names | DCP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1B (EC 3.-.-.-). | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.020160 (rank : 104) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.020094 (rank : 105) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.019889 (rank : 106) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MYOC_MOUSE
|
||||||
| NC score | 0.019815 (rank : 107) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O70624, O70289 | Gene names | Myoc, Tigr | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.019559 (rank : 108) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
SH24A_HUMAN
|
||||||
| NC score | 0.019438 (rank : 109) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.018930 (rank : 110) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.017087 (rank : 111) | θ value | 0.279714 (rank : 57) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.016976 (rank : 112) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.016903 (rank : 113) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.016596 (rank : 114) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.016015 (rank : 115) | θ value | 0.47712 (rank : 64) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.015921 (rank : 116) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
BMF_HUMAN
|
||||||
| NC score | 0.015746 (rank : 117) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96LC9, Q2M396, Q6NT30, Q6NT56, Q6P9F6, Q7Z7D4, Q7Z7D5, Q9H7K7 | Gene names | BMF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-modifying factor. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.013021 (rank : 118) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.012963 (rank : 119) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.012522 (rank : 120) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.012331 (rank : 121) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.011248 (rank : 122) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
NFKB2_HUMAN
|
||||||
| NC score | 0.010701 (rank : 123) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.010613 (rank : 124) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.010538 (rank : 125) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
CLC4F_HUMAN
|
||||||
| NC score | 0.010242 (rank : 126) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N1N0 | Gene names | CLEC4F, CLECSF13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13). | |||||
|
NFKB2_MOUSE
|
||||||
| NC score | 0.010125 (rank : 127) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.010055 (rank : 128) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.008937 (rank : 129) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
NARGL_MOUSE
|
||||||
| NC score | 0.006982 (rank : 130) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
FRS2_MOUSE
|
||||||
| NC score | 0.005342 (rank : 131) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C180 | Gene names | Frs2, Frs2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 2 (FGFR substrate 2) (Suc1-associated neurotrophic factor target 1) (SNT-1) (FGFR signaling adaptor SNT) (FRS2 alpha). | |||||
|
TOM1_MOUSE
|
||||||
| NC score | 0.004661 (rank : 132) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.003914 (rank : 133) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.002813 (rank : 134) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | 0.001191 (rank : 135) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
ZN148_MOUSE
|
||||||
| NC score | -0.002623 (rank : 136) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||