Please be patient as the page loads
|
HIF1A_MOUSE
|
||||||
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HIF1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993764 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 2.04462e-161 (rank : 3) | NC score | 0.977646 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 1.01473e-160 (rank : 4) | NC score | 0.978948 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 1.89707e-74 (rank : 5) | NC score | 0.936152 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 7.20884e-74 (rank : 6) | NC score | 0.935205 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 4.67263e-73 (rank : 7) | NC score | 0.933396 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 1.7756e-72 (rank : 8) | NC score | 0.932898 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 8.00737e-57 (rank : 9) | NC score | 0.907277 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 1.36585e-56 (rank : 10) | NC score | 0.907626 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 1.91031e-50 (rank : 11) | NC score | 0.908878 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 8.02596e-49 (rank : 12) | NC score | 0.909256 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 13) | NC score | 0.825732 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 1.0531e-32 (rank : 14) | NC score | 0.823646 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 15) | NC score | 0.802272 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 2.68423e-28 (rank : 16) | NC score | 0.827578 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 5.97985e-28 (rank : 17) | NC score | 0.798932 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 18) | NC score | 0.830453 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 19) | NC score | 0.788253 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 1.47289e-26 (rank : 20) | NC score | 0.776019 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.47289e-26 (rank : 21) | NC score | 0.762959 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 1.47289e-26 (rank : 22) | NC score | 0.781677 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 7.30988e-26 (rank : 23) | NC score | 0.781496 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 24) | NC score | 0.780485 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 25) | NC score | 0.675124 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 26) | NC score | 0.657315 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 27) | NC score | 0.566264 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 28) | NC score | 0.565268 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 29) | NC score | 0.621006 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 30) | NC score | 0.623619 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 31) | NC score | 0.566854 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 32) | NC score | 0.529454 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 33) | NC score | 0.340095 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 34) | NC score | 0.315634 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 35) | NC score | 0.313470 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 36) | NC score | 0.302693 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 37) | NC score | 0.309678 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 38) | NC score | 0.285995 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
TRI37_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 39) | NC score | 0.020704 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94972, Q7Z3E6, Q8IYF7, Q8WYF7 | Gene names | TRIM37, KIAA0898, MUL, POB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37 (Mulibrey nanism protein). | |||||
|
CJ078_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.042691 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XK3, Q5JT40, Q8WW47 | Gene names | C10orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.010714 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.036417 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.013946 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.047137 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.010194 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.022790 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
KI2L4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.007682 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99706, O14621, O14622, O14623, O14624, O43534, P78400, P78401, Q99559, Q99560, Q99561, Q99562, Q9UQJ7 | Gene names | KIR2DL4, CD158D, KIR103AS | |||
|
Domain Architecture |
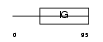
|
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL4 precursor (MHC class I NK cell receptor KIR103AS) (Killer cell inhibitory receptor 103AS) (KIR-103AS) (G9P) (CD158d antigen). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.031062 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | -0.001937 (rank : 105) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SHE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.008791 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VZ18, Q8TEQ5 | Gene names | SHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
SN1L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.000727 (rank : 101) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
SPAG5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.006179 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
ICAL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.012549 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.012662 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.009636 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PKHA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.009761 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
DDHD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.018310 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.008495 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
INT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.014491 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P4S8, Q80UQ7, Q91Z01, Q9CTF7 | Gene names | Ints1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.003334 (rank : 96) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.004629 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
PASK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.002286 (rank : 97) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
PCDAB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | -0.000380 (rank : 103) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.021188 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.008825 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
K1377_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.011015 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2H0, Q4G0U6 | Gene names | KIAA1377 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1377. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007077 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.008785 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.023932 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | -0.000946 (rank : 104) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
UT14A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.005165 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
ZN406_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | -0.003920 (rank : 107) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P243, Q3MIM5, Q6PJ01, Q75PJ7, Q75PJ9, Q86X64 | Gene names | ZNF406, KIAA1485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406 (Protein ZFAT). | |||||
|
AKTS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.008670 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B36, Q96BI4, Q96IK7, Q96NG2, Q9BWR5 | Gene names | AKT1S1, PRAS40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (40 kDa proline-rich AKT substrate). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.003759 (rank : 95) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.001497 (rank : 99) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.022269 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.004776 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
EOMES_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.002068 (rank : 98) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |
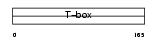
|
|||||
| Description | Eomesodermin homolog. | |||||
|
GAK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | -0.002172 (rank : 106) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.004300 (rank : 93) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.028686 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.028742 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.005579 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
INT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.012080 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N201, Q6NT70, Q6UX74, Q8WV40, Q96D36, Q9NTD1, Q9P2A8, Q9Y3W8 | Gene names | INTS1, KIAA1440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.001200 (rank : 100) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.000690 (rank : 102) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.018485 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.010343 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
TAGAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.003910 (rank : 94) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.021161 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.062725 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.081825 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.085639 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.092252 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.090184 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.072666 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.074353 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.065299 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.065290 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.075700 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.074094 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.068560 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.068444 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.068005 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.067973 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.065608 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.060977 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.993764 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.978948 (rank : 3) | θ value | 1.01473e-160 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.977646 (rank : 4) | θ value | 2.04462e-161 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.936152 (rank : 5) | θ value | 1.89707e-74 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.935205 (rank : 6) | θ value | 7.20884e-74 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.933396 (rank : 7) | θ value | 4.67263e-73 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.932898 (rank : 8) | θ value | 1.7756e-72 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.909256 (rank : 9) | θ value | 8.02596e-49 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.908878 (rank : 10) | θ value | 1.91031e-50 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.907626 (rank : 11) | θ value | 1.36585e-56 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.907277 (rank : 12) | θ value | 8.00737e-57 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.830453 (rank : 13) | θ value | 7.80994e-28 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.827578 (rank : 14) | θ value | 2.68423e-28 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.825732 (rank : 15) | θ value | 1.0531e-32 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.823646 (rank : 16) | θ value | 1.0531e-32 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.802272 (rank : 17) | θ value | 1.57365e-28 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.798932 (rank : 18) | θ value | 5.97985e-28 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.788253 (rank : 19) | θ value | 1.12775e-26 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.781677 (rank : 20) | θ value | 1.47289e-26 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.781496 (rank : 21) | θ value | 7.30988e-26 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.780485 (rank : 22) | θ value | 1.24688e-25 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.776019 (rank : 23) | θ value | 1.47289e-26 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.762959 (rank : 24) | θ value | 1.47289e-26 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.675124 (rank : 25) | θ value | 1.38178e-16 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.657315 (rank : 26) | θ value | 2.88119e-14 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.623619 (rank : 27) | θ value | 2.69671e-12 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.621006 (rank : 28) | θ value | 1.58096e-12 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.566854 (rank : 29) | θ value | 6.64225e-11 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.566264 (rank : 30) | θ value | 1.86753e-13 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.565268 (rank : 31) | θ value | 1.2105e-12 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.529454 (rank : 32) | θ value | 3.64472e-09 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.340095 (rank : 33) | θ value | 6.43352e-06 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.315634 (rank : 34) | θ value | 1.43324e-05 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.313470 (rank : 35) | θ value | 1.43324e-05 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.309678 (rank : 36) | θ value | 0.00134147 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.302693 (rank : 37) | θ value | 0.00020696 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.285995 (rank : 38) | θ value | 0.0113563 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.092252 (rank : 39) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.090184 (rank : 40) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.085639 (rank : 41) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.081825 (rank : 42) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.075700 (rank : 43) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.074353 (rank : 44) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.074094 (rank : 45) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.072666 (rank : 46) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.068560 (rank : 47) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.068444 (rank : 48) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.068005 (rank : 49) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.067973 (rank : 50) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.065608 (rank : 51) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.065299 (rank : 52) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.065290 (rank : 53) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.062725 (rank : 54) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.060977 (rank : 55) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.047137 (rank : 56) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
CJ078_HUMAN
|
||||||
| NC score | 0.042691 (rank : 57) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XK3, Q5JT40, Q8WW47 | Gene names | C10orf78 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.036417 (rank : 58) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.031062 (rank : 59) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.028742 (rank : 60) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.028686 (rank : 61) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.023932 (rank : 62) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.022790 (rank : 63) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.022269 (rank : 64) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.021188 (rank : 65) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.021161 (rank : 66) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRI37_HUMAN
|
||||||
| NC score | 0.020704 (rank : 67) | θ value | 0.0736092 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94972, Q7Z3E6, Q8IYF7, Q8WYF7 | Gene names | TRIM37, KIAA0898, MUL, POB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 37 (Mulibrey nanism protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.018485 (rank : 68) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
DDHD1_HUMAN
|
||||||
| NC score | 0.018310 (rank : 69) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEL9, Q8WVH3, Q96LL2, Q9C0F8 | Gene names | DDHD1, KIAA1705 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipase DDHD1 (EC 3.1.1.-) (DDHD domain protein 1) (Phosphatidic acid-preferring phospholipase A1 homolog) (PA-PLA1). | |||||
|
INT1_MOUSE
|
||||||
| NC score | 0.014491 (rank : 70) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P4S8, Q80UQ7, Q91Z01, Q9CTF7 | Gene names | Ints1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.013946 (rank : 71) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.012662 (rank : 72) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.012549 (rank : 73) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
INT1_HUMAN
|
||||||
| NC score | 0.012080 (rank : 74) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N201, Q6NT70, Q6UX74, Q8WV40, Q96D36, Q9NTD1, Q9P2A8, Q9Y3W8 | Gene names | INTS1, KIAA1440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 1 (Int1). | |||||
|
K1377_HUMAN
|
||||||
| NC score | 0.011015 (rank : 75) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2H0, Q4G0U6 | Gene names | KIAA1377 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1377. | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.010714 (rank : 76) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.010343 (rank : 77) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.010194 (rank : 78) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
PKHA4_MOUSE
|
||||||
| NC score | 0.009761 (rank : 79) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VC98, Q8R3N3 | Gene names | Plekha4, Pepp1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.009636 (rank : 80) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.008825 (rank : 81) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
SHE_HUMAN
|
||||||
| NC score | 0.008791 (rank : 82) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VZ18, Q8TEQ5 | Gene names | SHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.008785 (rank : 83) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
AKTS1_HUMAN
|
||||||
| NC score | 0.008670 (rank : 84) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B36, Q96BI4, Q96IK7, Q96NG2, Q9BWR5 | Gene names | AKT1S1, PRAS40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (40 kDa proline-rich AKT substrate). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.008495 (rank : 85) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
KI2L4_HUMAN
|
||||||
| NC score | 0.007682 (rank : 86) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99706, O14621, O14622, O14623, O14624, O43534, P78400, P78401, Q99559, Q99560, Q99561, Q99562, Q9UQJ7 | Gene names | KIR2DL4, CD158D, KIR103AS | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL4 precursor (MHC class I NK cell receptor KIR103AS) (Killer cell inhibitory receptor 103AS) (KIR-103AS) (G9P) (CD158d antigen). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.007077 (rank : 87) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SPAG5_MOUSE
|
||||||
| NC score | 0.006179 (rank : 88) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 612 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TME2, Q8CH61 | Gene names | Spag5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Mastrin) (Mitotic spindle-associated protein p126) (MAP126). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.005579 (rank : 89) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
UT14A_MOUSE
|
||||||
| NC score | 0.005165 (rank : 90) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.004776 (rank : 91) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.004629 (rank : 92) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.004300 (rank : 93) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
TAGAP_MOUSE
|
||||||
| NC score | 0.003910 (rank : 94) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2L9, Q920D6 | Gene names | Tagap, Tagap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell activation Rho GTPase-activating protein (T-cell activation GTPase-activating protein). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.003759 (rank : 95) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.003334 (rank : 96) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
PASK_MOUSE
|
||||||
| NC score | 0.002286 (rank : 97) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
EOMES_HUMAN
|
||||||
| NC score | 0.002068 (rank : 98) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.001497 (rank : 99) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.001200 (rank : 100) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
SN1L2_HUMAN
|
||||||
| NC score | 0.000727 (rank : 101) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.000690 (rank : 102) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
PCDAB_HUMAN
|
||||||
| NC score | -0.000380 (rank : 103) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | -0.000946 (rank : 104) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
STK10_MOUSE
|
||||||
| NC score | -0.001937 (rank : 105) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
GAK_HUMAN
|
||||||
| NC score | -0.002172 (rank : 106) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
ZN406_HUMAN
|
||||||
| NC score | -0.003920 (rank : 107) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P243, Q3MIM5, Q6PJ01, Q75PJ7, Q75PJ9, Q86X64 | Gene names | ZNF406, KIAA1485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406 (Protein ZFAT). | |||||