Please be patient as the page loads
|
EPAS1_HUMAN
|
||||||
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EPAS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994197 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 3.1544e-162 (rank : 3) | NC score | 0.976450 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 1.01473e-160 (rank : 4) | NC score | 0.978948 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 7.98882e-65 (rank : 5) | NC score | 0.924058 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 8.83269e-64 (rank : 6) | NC score | 0.922236 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 9.76566e-63 (rank : 7) | NC score | 0.921283 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 9.76566e-63 (rank : 8) | NC score | 0.924438 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 3.974e-56 (rank : 9) | NC score | 0.900651 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 6.77864e-56 (rank : 10) | NC score | 0.900592 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 4.70527e-49 (rank : 11) | NC score | 0.901675 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 12) | NC score | 0.900224 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 13) | NC score | 0.830571 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 1.37539e-32 (rank : 14) | NC score | 0.811382 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 15) | NC score | 0.828226 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 16) | NC score | 0.807894 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 17) | NC score | 0.799120 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 18) | NC score | 0.793388 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 19) | NC score | 0.794169 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 20) | NC score | 0.788852 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 6.38894e-30 (rank : 21) | NC score | 0.792996 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 22) | NC score | 0.774905 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 23) | NC score | 0.812940 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 24) | NC score | 0.816161 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 25) | NC score | 0.673081 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 26) | NC score | 0.656582 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 27) | NC score | 0.563213 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 28) | NC score | 0.562488 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 29) | NC score | 0.566111 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 30) | NC score | 0.614588 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 31) | NC score | 0.617030 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 32) | NC score | 0.529781 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 33) | NC score | 0.322580 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 34) | NC score | 0.325094 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 35) | NC score | 0.347110 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 36) | NC score | 0.309179 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 37) | NC score | 0.317250 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 38) | NC score | 0.293878 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
FANCJ_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.031788 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SXJ3, Q8BJQ8, Q8BKI6 | Gene names | Brip1, Bach1, Fancj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
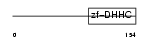
ZDHC5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 40) | NC score | 0.017624 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.021813 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
PASK_HUMAN
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.007607 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RG2, Q86XH6, Q99763, Q9UFR7 | Gene names | PASK, KIAA0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN) (hPASK). | |||||
|
UFO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.000282 (rank : 91) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00993 | Gene names | Axl, Ark, Ufo | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor UFO precursor (EC 2.7.10.1) (Adhesion-related kinase). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | -0.000662 (rank : 92) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.012981 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.006205 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
GBP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.008653 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01514 | Gene names | Gbp1, Gbp-1, Mag-1, Mpa1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (mGBP1) (mGBP-1) (Interferon-gamma-inducible protein MAG-1). | |||||
|
PASK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.005560 (rank : 79) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
RAIN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.013593 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U0S6, Q6GQW4 | Gene names | Rasip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.011391 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.032333 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
DPP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.006925 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TI2, Q6AI37, Q6UAL0, Q6ZMT2, Q6ZNJ5, Q8N2J7, Q8N3F5, Q8WXD8, Q96NT8, Q9BVR3 | Gene names | DPP9, DPRP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 9 (EC 3.4.14.5) (Dipeptidyl peptidase IX) (DP9) (Dipeptidyl peptidase-like protein 9) (DPLP9) (Dipeptidyl peptidase IV-related protein 2) (DPRP-2). | |||||
|
FBX42_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.009538 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3S6, Q5TEU8, Q86XI0, Q8N3N4, Q8N5F8, Q9BRM0, Q9P2L4 | Gene names | FBXO42, FBX42, KIAA1332 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 42. | |||||
|
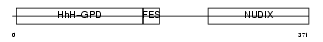
MUTYH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.015617 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UIF7, Q15830, Q9UBP2, Q9UBS7, Q9UIF4, Q9UIF5, Q9UIF6 | Gene names | MUTYH, MYH | |||
|
Domain Architecture |
|
|||||
| Description | A/G-specific adenine DNA glycosylase (EC 3.2.2.-) (MutY homolog) (hMYH). | |||||
|
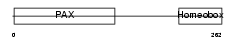
PAX6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.003236 (rank : 85) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26367, Q6N006, Q99413 | Gene names | PAX6, AN2 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-6 (Oculorhombin) (Aniridia type II protein). | |||||
|
PAX6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.003236 (rank : 86) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63015, P32117, P70601, Q62222, Q64037, Q8CEI5, Q8VDB5, Q921Q8 | Gene names | Pax6, Pax-6, Sey | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-6 (Oculorhombin). | |||||
|
SPG16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.003005 (rank : 87) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K450, Q8C450, Q9CPU8, Q9D522 | Gene names | Spag16, Pf20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 16 protein (PF20 protein homolog). | |||||
|
WASF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.015760 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ABRA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.015731 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.007396 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
RX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.002768 (rank : 88) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.005263 (rank : 81) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.016478 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.011911 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.016209 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.009445 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
KLF8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | -0.003793 (rank : 94) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95600, Q59GV5, Q5HYQ5, Q5JXP7, Q6MZJ7, Q9UGC4 | Gene names | KLF8, BKLF3, ZNF741 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 8 (Basic krueppel-like factor 3) (Zinc finger protein 741). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.010407 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | -0.002824 (rank : 93) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.000526 (rank : 90) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.007238 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
ESAM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.003507 (rank : 84) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
PCGF6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.004316 (rank : 83) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYE7, Q5SYD1, Q5SYD6, Q96ID9, Q96SJ1 | Gene names | PCGF6, MBLR, RNF134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb group RING finger protein 6 (RING finger protein 134) (Mel18 and Bmi1-like RING finger). | |||||
|
RHBD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.005484 (rank : 80) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75783, Q9NQ85 | Gene names | RHBDL1, RHBDL | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 1 (EC 3.4.21.105) (RRP) (Rhomboid-like protein 1). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.005044 (rank : 82) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.002647 (rank : 89) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.061536 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.081031 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.084934 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.091860 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.089845 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.075181 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.077147 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.067531 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.067571 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.050982 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.078977 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.077619 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.071834 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.071817 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.071060 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.071028 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.068232 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.063441 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.994197 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.978948 (rank : 3) | θ value | 1.01473e-160 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.976450 (rank : 4) | θ value | 3.1544e-162 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.924438 (rank : 5) | θ value | 9.76566e-63 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.924058 (rank : 6) | θ value | 7.98882e-65 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.922236 (rank : 7) | θ value | 8.83269e-64 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.921283 (rank : 8) | θ value | 9.76566e-63 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.901675 (rank : 9) | θ value | 4.70527e-49 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.900651 (rank : 10) | θ value | 3.974e-56 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.900592 (rank : 11) | θ value | 6.77864e-56 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.900224 (rank : 12) | θ value | 2.3352e-48 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.830571 (rank : 13) | θ value | 1.0531e-32 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.828226 (rank : 14) | θ value | 1.79631e-32 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.816161 (rank : 15) | θ value | 1.6852e-22 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.812940 (rank : 16) | θ value | 1.99067e-23 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.811382 (rank : 17) | θ value | 1.37539e-32 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.807894 (rank : 18) | θ value | 1.16434e-31 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.799120 (rank : 19) | θ value | 2.59387e-31 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.794169 (rank : 20) | θ value | 1.28732e-30 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.793388 (rank : 21) | θ value | 9.8567e-31 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.792996 (rank : 22) | θ value | 6.38894e-30 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.788852 (rank : 23) | θ value | 3.74554e-30 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.774905 (rank : 24) | θ value | 1.08979e-29 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.673081 (rank : 25) | θ value | 3.63628e-17 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.656582 (rank : 26) | θ value | 4.02038e-16 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.617030 (rank : 27) | θ value | 8.67504e-11 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.614588 (rank : 28) | θ value | 3.89403e-11 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.566111 (rank : 29) | θ value | 2.28291e-11 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.563213 (rank : 30) | θ value | 7.09661e-13 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.562488 (rank : 31) | θ value | 2.0648e-12 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.529781 (rank : 32) | θ value | 7.34386e-10 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.347110 (rank : 33) | θ value | 3.19293e-05 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.325094 (rank : 34) | θ value | 8.40245e-06 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.322580 (rank : 35) | θ value | 6.43352e-06 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.317250 (rank : 36) | θ value | 0.00134147 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.309179 (rank : 37) | θ value | 0.00020696 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.293878 (rank : 38) | θ value | 0.00390308 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.091860 (rank : 39) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.089845 (rank : 40) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.084934 (rank : 41) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.081031 (rank : 42) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.078977 (rank : 43) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.077619 (rank : 44) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.077147 (rank : 45) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.075181 (rank : 46) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.071834 (rank : 47) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.071817 (rank : 48) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.071060 (rank : 49) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.071028 (rank : 50) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.068232 (rank : 51) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.067571 (rank : 52) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.067531 (rank : 53) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.063441 (rank : 54) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.061536 (rank : 55) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.050982 (rank : 56) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.032333 (rank : 57) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
FANCJ_MOUSE
|
||||||
| NC score | 0.031788 (rank : 58) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SXJ3, Q8BJQ8, Q8BKI6 | Gene names | Brip1, Bach1, Fancj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein homolog (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.021813 (rank : 59) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
ZDHC5_HUMAN
|
||||||
| NC score | 0.017624 (rank : 60) | θ value | 0.0563607 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.016478 (rank : 61) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.016209 (rank : 62) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
WASF2_HUMAN
|
||||||
| NC score | 0.015760 (rank : 63) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6W5, O60794, Q9UDY7 | Gene names | WASF2, WAVE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2) (Verprolin homology domain-containing protein 2). | |||||
|
ABRA_HUMAN
|
||||||
| NC score | 0.015731 (rank : 64) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
MUTYH_HUMAN
|
||||||
| NC score | 0.015617 (rank : 65) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UIF7, Q15830, Q9UBP2, Q9UBS7, Q9UIF4, Q9UIF5, Q9UIF6 | Gene names | MUTYH, MYH | |||
|
Domain Architecture |

|
|||||
| Description | A/G-specific adenine DNA glycosylase (EC 3.2.2.-) (MutY homolog) (hMYH). | |||||
|
RAIN_MOUSE
|
||||||
| NC score | 0.013593 (rank : 66) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U0S6, Q6GQW4 | Gene names | Rasip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.012981 (rank : 67) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.011911 (rank : 68) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.011391 (rank : 69) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.010407 (rank : 70) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
FBX42_HUMAN
|
||||||
| NC score | 0.009538 (rank : 71) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3S6, Q5TEU8, Q86XI0, Q8N3N4, Q8N5F8, Q9BRM0, Q9P2L4 | Gene names | FBXO42, FBX42, KIAA1332 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 42. | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.009445 (rank : 72) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
GBP1_MOUSE
|
||||||
| NC score | 0.008653 (rank : 73) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01514 | Gene names | Gbp1, Gbp-1, Mag-1, Mpa1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (mGBP1) (mGBP-1) (Interferon-gamma-inducible protein MAG-1). | |||||
|
PASK_HUMAN
|
||||||
| NC score | 0.007607 (rank : 74) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RG2, Q86XH6, Q99763, Q9UFR7 | Gene names | PASK, KIAA0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN) (hPASK). | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.007396 (rank : 75) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.007238 (rank : 76) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DPP9_HUMAN
|
||||||
| NC score | 0.006925 (rank : 77) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TI2, Q6AI37, Q6UAL0, Q6ZMT2, Q6ZNJ5, Q8N2J7, Q8N3F5, Q8WXD8, Q96NT8, Q9BVR3 | Gene names | DPP9, DPRP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 9 (EC 3.4.14.5) (Dipeptidyl peptidase IX) (DP9) (Dipeptidyl peptidase-like protein 9) (DPLP9) (Dipeptidyl peptidase IV-related protein 2) (DPRP-2). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.006205 (rank : 78) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
PASK_MOUSE
|
||||||
| NC score | 0.005560 (rank : 79) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
RHBD1_HUMAN
|
||||||
| NC score | 0.005484 (rank : 80) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75783, Q9NQ85 | Gene names | RHBDL1, RHBDL | |||
|
Domain Architecture |

|
|||||
| Description | Rhomboid-related protein 1 (EC 3.4.21.105) (RRP) (Rhomboid-like protein 1). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.005263 (rank : 81) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.005044 (rank : 82) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
PCGF6_HUMAN
|
||||||
| NC score | 0.004316 (rank : 83) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYE7, Q5SYD1, Q5SYD6, Q96ID9, Q96SJ1 | Gene names | PCGF6, MBLR, RNF134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb group RING finger protein 6 (RING finger protein 134) (Mel18 and Bmi1-like RING finger). | |||||
|
ESAM_HUMAN
|
||||||
| NC score | 0.003507 (rank : 84) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96AP7, Q96T50 | Gene names | ESAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-selective adhesion molecule precursor. | |||||
|
PAX6_HUMAN
|
||||||
| NC score | 0.003236 (rank : 85) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26367, Q6N006, Q99413 | Gene names | PAX6, AN2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-6 (Oculorhombin) (Aniridia type II protein). | |||||
|
PAX6_MOUSE
|
||||||
| NC score | 0.003236 (rank : 86) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63015, P32117, P70601, Q62222, Q64037, Q8CEI5, Q8VDB5, Q921Q8 | Gene names | Pax6, Pax-6, Sey | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-6 (Oculorhombin). | |||||
|
SPG16_MOUSE
|
||||||
| NC score | 0.003005 (rank : 87) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K450, Q8C450, Q9CPU8, Q9D522 | Gene names | Spag16, Pf20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 16 protein (PF20 protein homolog). | |||||
|
RX_HUMAN
|
||||||
| NC score | 0.002768 (rank : 88) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.002647 (rank : 89) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.000526 (rank : 90) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
UFO_MOUSE
|
||||||
| NC score | 0.000282 (rank : 91) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00993 | Gene names | Axl, Ark, Ufo | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor UFO precursor (EC 2.7.10.1) (Adhesion-related kinase). | |||||
|
STK10_MOUSE
|
||||||
| NC score | -0.000662 (rank : 92) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SLK_HUMAN
|
||||||
| NC score | -0.002824 (rank : 93) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
KLF8_HUMAN
|
||||||
| NC score | -0.003793 (rank : 94) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95600, Q59GV5, Q5HYQ5, Q5JXP7, Q6MZJ7, Q9UGC4 | Gene names | KLF8, BKLF3, ZNF741 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 8 (Basic krueppel-like factor 3) (Zinc finger protein 741). | |||||