Please be patient as the page loads
|
ARNT2_HUMAN
|
||||||
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ARNT2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997952 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.988031 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.973755 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 1.49618e-87 (rank : 5) | NC score | 0.941435 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 4.81303e-86 (rank : 6) | NC score | 0.939666 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 8.91499e-32 (rank : 7) | NC score | 0.794139 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 8) | NC score | 0.794169 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 9) | NC score | 0.780297 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 10) | NC score | 0.781496 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 2.77775e-25 (rank : 11) | NC score | 0.748487 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 12) | NC score | 0.792269 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 13) | NC score | 0.792793 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 14) | NC score | 0.753431 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 1.80048e-24 (rank : 15) | NC score | 0.752354 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 2.35151e-24 (rank : 16) | NC score | 0.796005 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 1.16704e-23 (rank : 17) | NC score | 0.747697 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 18) | NC score | 0.795088 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 19) | NC score | 0.709201 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 20) | NC score | 0.702335 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 21) | NC score | 0.574792 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 22) | NC score | 0.610070 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 23) | NC score | 0.606035 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 24) | NC score | 0.687367 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
PER2_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 25) | NC score | 0.480668 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 26) | NC score | 0.450535 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 27) | NC score | 0.536153 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 28) | NC score | 0.682830 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 29) | NC score | 0.454048 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 30) | NC score | 0.472369 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 31) | NC score | 0.469361 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 32) | NC score | 0.665594 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 33) | NC score | 0.488706 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 34) | NC score | 0.650743 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 35) | NC score | 0.523514 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 36) | NC score | 0.524576 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 37) | NC score | 0.506727 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 38) | NC score | 0.258175 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 39) | NC score | 0.266239 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 40) | NC score | 0.266360 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 41) | NC score | 0.249221 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 42) | NC score | 0.187568 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 43) | NC score | 0.231516 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 44) | NC score | 0.228539 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.181403 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 46) | NC score | 0.239068 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 47) | NC score | 0.486745 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 48) | NC score | 0.231643 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 49) | NC score | 0.231682 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 50) | NC score | 0.233568 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 51) | NC score | 0.226931 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 52) | NC score | 0.199901 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 53) | NC score | 0.206920 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 54) | NC score | 0.023051 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 0.21417 (rank : 55) | NC score | 0.175265 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.127967 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.124522 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYC_MOUSE
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.122023 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 59) | NC score | 0.123502 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 60) | NC score | 0.152408 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ZN485_HUMAN
|
||||||
| θ value | 0.365318 (rank : 61) | NC score | -0.001385 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NCK3, Q96CL0 | Gene names | ZNF485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 485. | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.032856 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.035257 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
MAX_HUMAN
|
||||||
| θ value | 0.47712 (rank : 64) | NC score | 0.164592 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 65) | NC score | 0.183829 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
BACH_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.039160 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V12, Q59HQ2 | Gene names | Acot7, Bach | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 67) | NC score | 0.121606 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.004750 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.012311 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.123886 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.024133 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.133967 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.120513 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.120346 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
EPHA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | -0.001445 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.124403 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
PLCD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.015555 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3B1 | Gene names | Plcd1, Plcd | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.126356 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.124663 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
CASL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.013827 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |
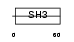
|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.036191 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
UFO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | -0.002788 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30530 | Gene names | AXL, UFO | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor UFO precursor (EC 2.7.10.1) (AXL oncogene). | |||||
|
FHOD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.019206 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.018729 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.072784 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.015726 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.020886 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
EPIPL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.014150 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
GAK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | -0.003192 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
MLX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.160457 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
ZN228_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | -0.003024 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJU3, Q9HCA7 | Gene names | ZNF228 | |||
|
Domain Architecture |
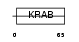
|
|||||
| Description | Zinc finger protein 228. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.020134 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCNT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.008565 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.008786 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.007971 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
MLX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.155168 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
NEDD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.003201 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33215, Q6NXV3, Q8BN12, Q8BN86, Q8BQL9, Q9CWK2 | Gene names | Nedd1, Nedd-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NEDD1 (Neural precursor cell expressed developmentally down- regulated protein 1). | |||||
|
PDE8A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.006986 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.024110 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.022758 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
RHES_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.003081 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63032, Q9WVD3 | Gene names | Rasd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein Rhes (Ras homolog enriched in striatum). | |||||
|
APOA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | -0.000885 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
CO9A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.001206 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14055 | Gene names | COL9A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
HES4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.094545 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.052844 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
PDE8A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.006031 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
RHES_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.002345 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96D21, O95520, Q5THY8 | Gene names | RASD2, TEM2 | |||
|
Domain Architecture |
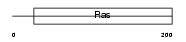
|
|||||
| Description | GTP-binding protein Rhes (Ras homolog enriched in striatum) (Tumor endothelial marker 2). | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.004967 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
ZNF7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | -0.003581 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17097, P17015 | Gene names | ZNF7, KOX4 | |||
|
Domain Architecture |
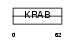
|
|||||
| Description | Zinc finger protein 7 (Zinc finger protein KOX4) (Zinc finger protein HF.16). | |||||
|
HES1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.073454 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.073803 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.064946 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.093902 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HES7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.061423 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.059284 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050407 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.061011 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.058052 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.050549 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.076119 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.997952 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.988031 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.973755 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.941435 (rank : 5) | θ value | 1.49618e-87 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.939666 (rank : 6) | θ value | 4.81303e-86 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.796005 (rank : 7) | θ value | 2.35151e-24 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.795088 (rank : 8) | θ value | 1.99067e-23 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.794169 (rank : 9) | θ value | 1.28732e-30 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.794139 (rank : 10) | θ value | 8.91499e-32 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.792793 (rank : 11) | θ value | 4.73814e-25 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.792269 (rank : 12) | θ value | 3.62785e-25 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.781496 (rank : 13) | θ value | 7.30988e-26 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.780297 (rank : 14) | θ value | 4.28545e-26 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.753431 (rank : 15) | θ value | 6.18819e-25 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.752354 (rank : 16) | θ value | 1.80048e-24 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.748487 (rank : 17) | θ value | 2.77775e-25 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.747697 (rank : 18) | θ value | 1.16704e-23 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.709201 (rank : 19) | θ value | 1.92812e-18 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.702335 (rank : 20) | θ value | 4.02038e-16 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.687367 (rank : 21) | θ value | 2.43908e-13 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.682830 (rank : 22) | θ value | 2.69671e-12 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.665594 (rank : 23) | θ value | 2.52405e-10 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.650743 (rank : 24) | θ value | 1.06045e-08 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.610070 (rank : 25) | θ value | 7.58209e-15 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.606035 (rank : 26) | θ value | 1.29331e-14 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.574792 (rank : 27) | θ value | 5.8054e-15 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.536153 (rank : 28) | θ value | 2.69671e-12 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.524576 (rank : 29) | θ value | 4.0297e-08 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.523514 (rank : 30) | θ value | 2.36244e-08 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.506727 (rank : 31) | θ value | 2.44474e-05 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.488706 (rank : 32) | θ value | 3.29651e-10 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.486745 (rank : 33) | θ value | 0.00869519 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.480668 (rank : 34) | θ value | 1.2105e-12 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.472369 (rank : 35) | θ value | 1.33837e-11 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.469361 (rank : 36) | θ value | 1.33837e-11 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.454048 (rank : 37) | θ value | 6.00763e-12 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.450535 (rank : 38) | θ value | 1.58096e-12 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.266360 (rank : 39) | θ value | 0.000786445 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.266239 (rank : 40) | θ value | 0.000786445 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.258175 (rank : 41) | θ value | 0.000602161 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.249221 (rank : 42) | θ value | 0.00102713 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.239068 (rank : 43) | θ value | 0.00665767 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.233568 (rank : 44) | θ value | 0.0113563 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.231682 (rank : 45) | θ value | 0.00869519 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.231643 (rank : 46) | θ value | 0.00869519 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.231516 (rank : 47) | θ value | 0.00298849 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.228539 (rank : 48) | θ value | 0.00298849 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.226931 (rank : 49) | θ value | 0.0431538 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.206920 (rank : 50) | θ value | 0.0736092 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.199901 (rank : 51) | θ value | 0.0736092 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.187568 (rank : 52) | θ value | 0.00175202 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.183829 (rank : 53) | θ value | 0.47712 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.181403 (rank : 54) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.175265 (rank : 55) | θ value | 0.21417 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.164592 (rank : 56) | θ value | 0.47712 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.160457 (rank : 57) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.155168 (rank : 58) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.152408 (rank : 59) | θ value | 0.279714 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.133967 (rank : 60) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.127967 (rank : 61) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.126356 (rank : 62) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.124663 (rank : 63) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.124522 (rank : 64) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.124403 (rank : 65) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.123886 (rank : 66) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.123502 (rank : 67) | θ value | 0.279714 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.122023 (rank : 68) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.121606 (rank : 69) | θ value | 0.62314 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.120513 (rank : 70) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.120346 (rank : 71) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.094545 (rank : 72) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.093902 (rank : 73) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.076119 (rank : 74) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.073803 (rank : 75) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.073454 (rank : 76) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.072784 (rank : 77) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.064946 (rank : 78) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.061423 (rank : 79) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.061011 (rank : 80) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.059284 (rank : 81) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.058052 (rank : 82) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.052844 (rank : 83) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.050549 (rank : 84) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.050407 (rank : 85) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
BACH_MOUSE
|
||||||
| NC score | 0.039160 (rank : 86) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V12, Q59HQ2 | Gene names | Acot7, Bach | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic acyl coenzyme A thioester hydrolase (EC 3.1.2.2) (Long chain acyl-CoA thioester hydrolase) (CTE-II) (CTE-IIa) (Brain acyl-CoA hydrolase) (Acyl-CoA thioesterase 7). | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.036191 (rank : 87) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.035257 (rank : 88) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.032856 (rank : 89) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.024133 (rank : 90) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.024110 (rank : 91) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.023051 (rank : 92) | θ value | 0.0961366 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.022758 (rank : 93) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.020886 (rank : 94) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.020134 (rank : 95) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
FHOD1_HUMAN
|
||||||
| NC score | 0.019206 (rank : 96) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.018729 (rank : 97) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.015726 (rank : 98) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
PLCD1_MOUSE
|
||||||
| NC score | 0.015555 (rank : 99) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3B1 | Gene names | Plcd1, Plcd | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
EPIPL_HUMAN
|
||||||
| NC score | 0.014150 (rank : 100) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
CASL_HUMAN
|
||||||
| NC score | 0.013827 (rank : 101) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14511 | Gene names | NEDD9, CASL | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of filamentation 1 (HEF1) (CRK-associated substrate-related protein) (CAS-L) (CasL) (p105) (Protein NEDD9) (NY-REN-12 antigen). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.012311 (rank : 102) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.008786 (rank : 103) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CCNT1_MOUSE
|
||||||
| NC score | 0.008565 (rank : 104) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.007971 (rank : 105) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
PDE8A_MOUSE
|
||||||
| NC score | 0.006986 (rank : 106) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88502 | Gene names | Pde8a, Pde8 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17) (MMPDE8). | |||||
|
PDE8A_HUMAN
|
||||||
| NC score | 0.006031 (rank : 107) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60658, Q969I1, Q96PC9, Q96PD0, Q96PD1, Q9UMB7 | Gene names | PDE8A | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A (EC 3.1.4.17). | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.004967 (rank : 108) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.004750 (rank : 109) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
NEDD1_MOUSE
|
||||||
| NC score | 0.003201 (rank : 110) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33215, Q6NXV3, Q8BN12, Q8BN86, Q8BQL9, Q9CWK2 | Gene names | Nedd1, Nedd-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NEDD1 (Neural precursor cell expressed developmentally down- regulated protein 1). | |||||
|
RHES_MOUSE
|
||||||
| NC score | 0.003081 (rank : 111) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63032, Q9WVD3 | Gene names | Rasd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein Rhes (Ras homolog enriched in striatum). | |||||
|
RHES_HUMAN
|
||||||
| NC score | 0.002345 (rank : 112) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96D21, O95520, Q5THY8 | Gene names | RASD2, TEM2 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Rhes (Ras homolog enriched in striatum) (Tumor endothelial marker 2). | |||||
|
CO9A2_HUMAN
|
||||||
| NC score | 0.001206 (rank : 113) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14055 | Gene names | COL9A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
APOA_HUMAN
|
||||||
| NC score | -0.000885 (rank : 114) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
ZN485_HUMAN
|
||||||
| NC score | -0.001385 (rank : 115) | θ value | 0.365318 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NCK3, Q96CL0 | Gene names | ZNF485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 485. | |||||
|
EPHA1_HUMAN
|
||||||
| NC score | -0.001445 (rank : 116) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
UFO_HUMAN
|
||||||
| NC score | -0.002788 (rank : 117) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30530 | Gene names | AXL, UFO | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor UFO precursor (EC 2.7.10.1) (AXL oncogene). | |||||
|
ZN228_HUMAN
|
||||||
| NC score | -0.003024 (rank : 118) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UJU3, Q9HCA7 | Gene names | ZNF228 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 228. | |||||
|
GAK_HUMAN
|
||||||
| NC score | -0.003192 (rank : 119) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
ZNF7_HUMAN
|
||||||
| NC score | -0.003581 (rank : 120) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17097, P17015 | Gene names | ZNF7, KOX4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 7 (Zinc finger protein KOX4) (Zinc finger protein HF.16). | |||||