Please be patient as the page loads
|
MAD4_MOUSE
|
||||||
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MAD4_MOUSE
|
||||||
| θ value | 8.75112e-96 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 1.22395e-89 (rank : 2) | NC score | 0.974956 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 5.39604e-37 (rank : 3) | NC score | 0.919926 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 4) | NC score | 0.919669 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 5) | NC score | 0.914988 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 2.77131e-33 (rank : 6) | NC score | 0.919582 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 7) | NC score | 0.891616 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 8) | NC score | 0.892869 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 9) | NC score | 0.431531 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 10) | NC score | 0.413792 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
RPC5_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 11) | NC score | 0.125750 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVU0, Q9BWF7, Q9H8W8, Q9H907, Q9P276 | Gene names | POLR3E, KIAA1452 | |||
|
Domain Architecture |
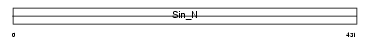
|
|||||
| Description | DNA-directed RNA polymerases III 80 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 5) (RPC5). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.313785 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.318226 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.276291 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.275535 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.275052 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.238531 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
USF1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.264506 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.265227 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.241276 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.027468 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.244848 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.080428 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.079485 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.121005 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.121685 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.035841 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
CING_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.030477 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.239953 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.038524 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.044693 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
NONO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.041108 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |
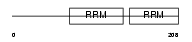
|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.041023 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.031837 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.026522 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.043394 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.200490 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.111977 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.136532 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
KCTD7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.029688 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
GBP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.024579 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P32455 | Gene names | GBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (HuGBP-1). | |||||
|
IF2M_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.028151 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |
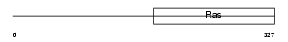
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.044942 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
ATF6B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.039827 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
NEUL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.022153 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYT8, Q9ULJ4 | Gene names | NLN, KIAA1226 | |||
|
Domain Architecture |

|
|||||
| Description | Neurolysin, mitochondrial precursor (EC 3.4.24.16) (Neurotensin endopeptidase) (Mitochondrial oligopeptidase M) (Microsomal endopeptidase) (MEP). | |||||
|
KCTD7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.022602 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MP8, Q8IVR0 | Gene names | KCTD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.018145 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
HES4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.086891 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.012614 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.013395 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.013659 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
ATF6B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.032942 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
BEGIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.027682 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.019813 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.006051 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.006177 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.016629 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.006803 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
BEGIN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.032818 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68EF6 | Gene names | Begain | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.105045 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.005010 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.005283 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
SNUT1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.012385 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.003914 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
ZFY19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.009346 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.050407 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.050680 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.062312 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.054888 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.091043 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.113384 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.105211 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
HES2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050747 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
MAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.224143 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.216249 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.144163 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.134485 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.054001 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.053711 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MYCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.112696 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
RPC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.065283 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZT4, Q8CI35, Q9DBD1 | Gene names | Polr3e, Sin | |||
|
Domain Architecture |
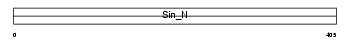
|
|||||
| Description | DNA-directed RNA polymerases III 80 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 5) (RPC5) (Sex-lethal interactor homolog). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.084293 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.152343 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.060629 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.154347 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.105930 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.105815 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.75112e-96 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.974956 (rank : 2) | θ value | 1.22395e-89 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.919926 (rank : 3) | θ value | 5.39604e-37 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.919669 (rank : 4) | θ value | 3.4976e-36 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.919582 (rank : 5) | θ value | 2.77131e-33 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.914988 (rank : 6) | θ value | 1.12514e-34 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.892869 (rank : 7) | θ value | 1.24688e-25 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.891616 (rank : 8) | θ value | 5.06226e-27 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.431531 (rank : 9) | θ value | 4.1701e-05 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.413792 (rank : 10) | θ value | 7.1131e-05 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.318226 (rank : 11) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.313785 (rank : 12) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.276291 (rank : 13) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.275535 (rank : 14) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.275052 (rank : 15) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.265227 (rank : 16) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.264506 (rank : 17) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.244848 (rank : 18) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.241276 (rank : 19) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.239953 (rank : 20) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.238531 (rank : 21) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.224143 (rank : 22) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.216249 (rank : 23) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.200490 (rank : 24) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.154347 (rank : 25) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.152343 (rank : 26) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.144163 (rank : 27) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.136532 (rank : 28) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.134485 (rank : 29) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
RPC5_HUMAN
|
||||||
| NC score | 0.125750 (rank : 30) | θ value | 0.000602161 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVU0, Q9BWF7, Q9H8W8, Q9H907, Q9P276 | Gene names | POLR3E, KIAA1452 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerases III 80 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 5) (RPC5). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.121685 (rank : 31) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.121005 (rank : 32) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.113384 (rank : 33) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.112696 (rank : 34) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.111977 (rank : 35) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.105930 (rank : 36) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.105815 (rank : 37) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.105211 (rank : 38) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.105045 (rank : 39) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.091043 (rank : 40) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.086891 (rank : 41) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.084293 (rank : 42) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.080428 (rank : 43) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.079485 (rank : 44) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
RPC5_MOUSE
|
||||||
| NC score | 0.065283 (rank : 45) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZT4, Q8CI35, Q9DBD1 | Gene names | Polr3e, Sin | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerases III 80 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 5) (RPC5) (Sex-lethal interactor homolog). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.062312 (rank : 46) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.060629 (rank : 47) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.054888 (rank : 48) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.054001 (rank : 49) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.053711 (rank : 50) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.050747 (rank : 51) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.050680 (rank : 52) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.050407 (rank : 53) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.044942 (rank : 54) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.044693 (rank : 55) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.043394 (rank : 56) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
NONO_HUMAN
|
||||||
| NC score | 0.041108 (rank : 57) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |

|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| NC score | 0.041023 (rank : 58) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
ATF6B_HUMAN
|
||||||
| NC score | 0.039827 (rank : 59) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.038524 (rank : 60) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.035841 (rank : 61) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
ATF6B_MOUSE
|
||||||
| NC score | 0.032942 (rank : 62) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
BEGIN_MOUSE
|
||||||
| NC score | 0.032818 (rank : 63) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68EF6 | Gene names | Begain | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.031837 (rank : 64) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.030477 (rank : 65) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
KCTD7_MOUSE
|
||||||
| NC score | 0.029688 (rank : 66) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
IF2M_MOUSE
|
||||||
| NC score | 0.028151 (rank : 67) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
BEGIN_HUMAN
|
||||||
| NC score | 0.027682 (rank : 68) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.027468 (rank : 69) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.026522 (rank : 70) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
GBP1_HUMAN
|
||||||
| NC score | 0.024579 (rank : 71) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P32455 | Gene names | GBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (HuGBP-1). | |||||
|
KCTD7_HUMAN
|
||||||
| NC score | 0.022602 (rank : 72) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96MP8, Q8IVR0 | Gene names | KCTD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
NEUL_HUMAN
|
||||||
| NC score | 0.022153 (rank : 73) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYT8, Q9ULJ4 | Gene names | NLN, KIAA1226 | |||
|
Domain Architecture |

|
|||||
| Description | Neurolysin, mitochondrial precursor (EC 3.4.24.16) (Neurotensin endopeptidase) (Mitochondrial oligopeptidase M) (Microsomal endopeptidase) (MEP). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.019813 (rank : 74) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.018145 (rank : 75) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.016629 (rank : 76) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.013659 (rank : 77) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.013395 (rank : 78) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.012614 (rank : 79) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
SNUT1_HUMAN
|
||||||
| NC score | 0.012385 (rank : 80) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
ZFY19_HUMAN
|
||||||
| NC score | 0.009346 (rank : 81) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96K21, Q86WC2, Q8WU96 | Gene names | ZFYVE19, MPFYVE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 19 (MLL partner containing FYVE domain). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.006803 (rank : 82) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
MINK1_MOUSE
|
||||||
| NC score | 0.006177 (rank : 83) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_HUMAN
|
||||||
| NC score | 0.006051 (rank : 84) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.005283 (rank : 85) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.005010 (rank : 86) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.003914 (rank : 87) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||