Please be patient as the page loads
|
ATF6A_HUMAN
|
||||||
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ATF6A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 174 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
ATF6B_HUMAN
|
||||||
| θ value | 6.96618e-77 (rank : 2) | NC score | 0.848346 (rank : 3) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
ATF6B_MOUSE
|
||||||
| θ value | 4.51533e-76 (rank : 3) | NC score | 0.849044 (rank : 2) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
ATF1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 4) | NC score | 0.422518 (rank : 6) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
ATF1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 5) | NC score | 0.424998 (rank : 5) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
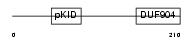
CREM_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 6) | NC score | 0.398879 (rank : 8) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |
|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREB3_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 7) | NC score | 0.426118 (rank : 4) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
CREB3_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 8) | NC score | 0.411427 (rank : 7) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREB1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 9) | NC score | 0.378600 (rank : 11) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
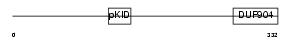
CREB1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 10) | NC score | 0.378628 (rank : 10) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |
|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREM_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 11) | NC score | 0.393416 (rank : 9) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
XBP1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 12) | NC score | 0.296997 (rank : 14) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
ZHANG_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 13) | NC score | 0.299649 (rank : 13) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NS37 | Gene names | ZF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor-binding transcription factor Zhangfei (HCF-binding transcription factor Zhangfei). | |||||
|
ZHANG_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 14) | NC score | 0.299946 (rank : 12) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZR3, Q91Y72 | Gene names | Zf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor-binding transcription factor Zhangfei (HCF-binding transcription factor Zhangfei) (Tyrosine kinase-associated leucine zipper protein LAZip). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.258411 (rank : 15) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 16) | NC score | 0.063126 (rank : 71) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
XBP1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 17) | NC score | 0.226589 (rank : 24) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 18) | NC score | 0.254698 (rank : 16) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.233581 (rank : 19) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.062262 (rank : 72) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 21) | NC score | 0.044651 (rank : 93) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ATF3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 22) | NC score | 0.237081 (rank : 17) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
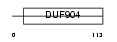
MAFK_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.170723 (rank : 32) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
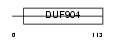
MAFK_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.170706 (rank : 33) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.129011 (rank : 49) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.021456 (rank : 155) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
CEBPE_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.116445 (rank : 51) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.180802 (rank : 30) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.183624 (rank : 29) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.047929 (rank : 88) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MAFG_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.170758 (rank : 31) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15525 | Gene names | MAFG | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G) (hMAF). | |||||
|
NRL_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.155094 (rank : 44) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
FOSB_MOUSE
|
||||||
| θ value | 0.125558 (rank : 33) | NC score | 0.158290 (rank : 40) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.164506 (rank : 36) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
MAFG_MOUSE
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.169713 (rank : 34) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
NFE2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.115747 (rank : 52) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
NRL_MOUSE
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.154934 (rank : 45) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54846 | Gene names | Nrl | |||
|
Domain Architecture |
|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.065683 (rank : 69) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.046986 (rank : 89) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.229744 (rank : 22) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.225625 (rank : 25) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.023871 (rank : 148) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.026968 (rank : 136) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PHC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.045296 (rank : 91) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.030442 (rank : 130) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.046818 (rank : 90) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.058749 (rank : 75) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.044387 (rank : 95) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
JUNB_HUMAN
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.234250 (rank : 18) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
JUNB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.231021 (rank : 21) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor jun-B. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.036966 (rank : 117) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.094944 (rank : 59) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
VPS45_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.055647 (rank : 79) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRW7, Q15715, Q9Y4Z6 | Gene names | VPS45A, VPS45 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 45 (h-VPS45) (hlVps45). | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.157385 (rank : 42) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
JUND_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.233394 (rank : 20) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.229268 (rank : 23) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.223413 (rank : 28) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 58) | NC score | 0.040615 (rank : 102) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.163951 (rank : 37) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.054604 (rank : 81) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.032526 (rank : 127) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.054779 (rank : 80) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
JUN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.225532 (rank : 27) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.225613 (rank : 26) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.037551 (rank : 114) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.121291 (rank : 50) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
VPS45_MOUSE
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.051812 (rank : 84) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97390, Q91VK9 | Gene names | Vps45a, Vps45 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 45 (mVps45). | |||||
|
BACH2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.073855 (rank : 67) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
CDC37_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.096411 (rank : 58) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
MAFB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.161410 (rank : 38) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
MAFF_HUMAN
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.158176 (rank : 41) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ULX9, Q9Y525 | Gene names | MAFF | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F) (U-Maf). | |||||
|
MAFF_MOUSE
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.160157 (rank : 39) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54791 | Gene names | Maff | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.044403 (rank : 94) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.027219 (rank : 134) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.010929 (rank : 179) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.027034 (rank : 135) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
BACH2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.070615 (rank : 68) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.065683 (rank : 70) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
CD248_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.019808 (rank : 158) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CEBPD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.090578 (rank : 60) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.145703 (rank : 46) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.038560 (rank : 113) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
MICA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.026466 (rank : 138) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.040924 (rank : 101) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.034584 (rank : 124) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
TEX15_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.041321 (rank : 99) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.022960 (rank : 149) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
BACH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.084318 (rank : 62) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.084275 (rank : 63) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.040979 (rank : 100) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.018263 (rank : 164) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.039660 (rank : 106) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.043084 (rank : 96) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
VDR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.008322 (rank : 184) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11473 | Gene names | VDR, NR1I1 | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
BATF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.155976 (rank : 43) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |
|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.017235 (rank : 167) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.023917 (rank : 147) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
G3BP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.018312 (rank : 163) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13283 | Gene names | G3BP, G3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.013349 (rank : 177) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.044942 (rank : 92) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.142888 (rank : 48) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.144207 (rank : 47) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.020282 (rank : 157) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
NDE1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.039921 (rank : 104) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.051296 (rank : 85) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.026213 (rank : 139) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.017263 (rank : 166) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
ZHX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.014971 (rank : 175) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.033903 (rank : 126) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.041752 (rank : 98) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
COVA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.042183 (rank : 97) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.018135 (rank : 165) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
FBXL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.025351 (rank : 143) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKT7, Q9P122 | Gene names | FBXL3, FBL3A, FBXL3A | |||
|
Domain Architecture |
|
|||||
| Description | F-box/LRR-repeat protein 3 (F-box/LRR-repeat protein 3A) (F-box and leucine-rich repeat protein 3A). | |||||
|
FBXL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.025382 (rank : 142) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4V4, Q80V32, Q8K1W0, Q9QXW1 | Gene names | Fbxl3, Fbl3a, Fbxl3a | |||
|
Domain Architecture |
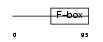
|
|||||
| Description | F-box/LRR-repeat protein 3 (F-box/LRR-repeat protein 3A) (F-box and leucine-rich repeat protein 3A). | |||||
|
KI18A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.015454 (rank : 173) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NI77, Q86VS5, Q9H0F3 | Gene names | KIF18A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.032279 (rank : 128) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.019625 (rank : 159) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.037196 (rank : 115) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.024798 (rank : 146) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
THAP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.022218 (rank : 153) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |
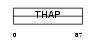
|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.039563 (rank : 107) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
BORG5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.021139 (rank : 156) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W92, Q9D8M1 | Gene names | Cdc42ep1, Borg5, Cep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5). | |||||
|
CING_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.038635 (rank : 112) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.019406 (rank : 160) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.022473 (rank : 151) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.038713 (rank : 111) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.016658 (rank : 170) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.031002 (rank : 129) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.036869 (rank : 119) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
MOR2A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.019156 (rank : 162) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.035001 (rank : 123) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYLK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.003458 (rank : 187) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
P73_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.015925 (rank : 172) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
PLVAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.034024 (rank : 125) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BX97, Q86VP0, Q8N8Y0, Q8ND68, Q8TER8, Q9BZD5 | Gene names | PLVAP, FELS, PV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plasmalemma vesicle-associated protein (Plasmalemma vesicle protein 1) (PV-1) (Fenestrated endothelial-linked structure protein). | |||||
|
POF1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.036117 (rank : 121) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.035538 (rank : 122) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ATF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.050454 (rank : 86) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.025392 (rank : 141) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
BATF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.166286 (rank : 35) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |
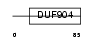
|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.025252 (rank : 144) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.037162 (rank : 116) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
COVA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.039414 (rank : 108) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |
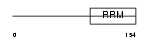
|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.038721 (rank : 110) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DAAM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.021503 (rank : 154) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.039889 (rank : 105) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.039163 (rank : 109) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
K1377_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.027469 (rank : 133) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2H0, Q4G0U6 | Gene names | KIAA1377 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1377. | |||||
|
L2GL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.009851 (rank : 181) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TJ91, Q6YP20, Q8K1X0 | Gene names | Llgl2, Llglh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(2) giant larvae protein homolog 2 (Lethal giant larvae-like protein 2). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.027936 (rank : 131) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
LRC59_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.009391 (rank : 182) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
MCA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.019291 (rank : 161) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12904, Q6FG28, Q96CQ9 | Gene names | SCYE1, EMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.025843 (rank : 140) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.036956 (rank : 118) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.026930 (rank : 137) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.053811 (rank : 82) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.008879 (rank : 183) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.022870 (rank : 150) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CING_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.040206 (rank : 103) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
FHOD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.014154 (rank : 176) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GEMI5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.006470 (rank : 185) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.010134 (rank : 180) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.022434 (rank : 152) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
HOME2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.027784 (rank : 132) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NSB8, O95269, O95349, Q9NSB6, Q9NSB7, Q9UNT7 | Gene names | HOMER2 | |||
|
Domain Architecture |
|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin). | |||||
|
MRCKG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.015364 (rank : 174) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.011256 (rank : 178) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NF2L3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.096959 (rank : 57) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.025018 (rank : 145) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.036477 (rank : 120) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
OTU6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.016277 (rank : 171) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L8S5 | Gene names | OTUD6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 6A. | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.050086 (rank : 87) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.006150 (rank : 186) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
VP33B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.017033 (rank : 168) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H267, Q96K14, Q9NRP6, Q9NSF3 | Gene names | VPS33B | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 33B (hVPS33B). | |||||
|
VP33B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.017009 (rank : 169) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59016 | Gene names | Vps33b | |||
|
Domain Architecture |
|
|||||
| Description | Vacuolar protein sorting-associated protein 33B. | |||||
|
ZN509_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | -0.002774 (rank : 188) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ATF5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.056399 (rank : 77) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
CEBPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.061615 (rank : 74) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.061754 (rank : 73) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.082607 (rank : 64) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CEBPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.076148 (rank : 65) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
CEBPD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.074876 (rank : 66) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
CEBPG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.057348 (rank : 76) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |
|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
CEBPG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.056291 (rank : 78) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
FOS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.111622 (rank : 53) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.110279 (rank : 54) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.100263 (rank : 55) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.097703 (rank : 56) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.086613 (rank : 61) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.052509 (rank : 83) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 174 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
ATF6B_MOUSE
|
||||||
| NC score | 0.849044 (rank : 2) | θ value | 4.51533e-76 (rank : 3) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
ATF6B_HUMAN
|
||||||
| NC score | 0.848346 (rank : 3) | θ value | 6.96618e-77 (rank : 2) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
CREB3_MOUSE
|
||||||
| NC score | 0.426118 (rank : 4) | θ value | 1.43324e-05 (rank : 7) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
ATF1_HUMAN
|
||||||
| NC score | 0.424998 (rank : 5) | θ value | 4.45536e-07 (rank : 5) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF1_MOUSE
|
||||||
| NC score | 0.422518 (rank : 6) | θ value | 2.61198e-07 (rank : 4) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
CREB3_HUMAN
|
||||||
| NC score | 0.411427 (rank : 7) | θ value | 1.87187e-05 (rank : 8) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREM_HUMAN
|
||||||
| NC score | 0.398879 (rank : 8) | θ value | 6.43352e-06 (rank : 6) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_MOUSE
|
||||||
| NC score | 0.393416 (rank : 9) | θ value | 7.1131e-05 (rank : 11) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREB1_MOUSE
|
||||||
| NC score | 0.378628 (rank : 10) | θ value | 7.1131e-05 (rank : 10) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_HUMAN
|
||||||
| NC score | 0.378600 (rank : 11) | θ value | 7.1131e-05 (rank : 9) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
ZHANG_MOUSE
|
||||||
| NC score | 0.299946 (rank : 12) | θ value | 0.000786445 (rank : 14) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZR3, Q91Y72 | Gene names | Zf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor-binding transcription factor Zhangfei (HCF-binding transcription factor Zhangfei) (Tyrosine kinase-associated leucine zipper protein LAZip). | |||||
|
ZHANG_HUMAN
|
||||||
| NC score | 0.299649 (rank : 13) | θ value | 0.000786445 (rank : 13) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NS37 | Gene names | ZF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor-binding transcription factor Zhangfei (HCF-binding transcription factor Zhangfei). | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.296997 (rank : 14) | θ value | 7.1131e-05 (rank : 12) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.258411 (rank : 15) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.254698 (rank : 16) | θ value | 0.00665767 (rank : 18) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.237081 (rank : 17) | θ value | 0.0193708 (rank : 22) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
JUNB_HUMAN
|
||||||
| NC score | 0.234250 (rank : 18) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.233581 (rank : 19) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.233394 (rank : 20) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUNB_MOUSE
|
||||||
| NC score | 0.231021 (rank : 21) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.229744 (rank : 22) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.229268 (rank : 23) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
XBP1_HUMAN
|
||||||
| NC score | 0.226589 (rank : 24) | θ value | 0.00175202 (rank : 17) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.225625 (rank : 25) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
JUN_MOUSE
|
||||||
| NC score | 0.225613 (rank : 26) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUN_HUMAN
|
||||||
| NC score | 0.225532 (rank : 27) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.223413 (rank : 28) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.183624 (rank : 29) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.180802 (rank : 30) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
MAFG_HUMAN
|
||||||
| NC score | 0.170758 (rank : 31) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15525 | Gene names | MAFG | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G) (hMAF). | |||||
|
MAFK_HUMAN
|
||||||
| NC score | 0.170723 (rank : 32) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O60675 | Gene names | MAFK | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFK_MOUSE
|
||||||
| NC score | 0.170706 (rank : 33) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61827, Q60600 | Gene names | Mafk, Nfe2u | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafK (Erythroid transcription factor NF-E2 p18 subunit). | |||||
|
MAFG_MOUSE
|
||||||
| NC score | 0.169713 (rank : 34) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54790 | Gene names | Mafg | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafG (V-maf musculoaponeurotic fibrosarcoma oncogene homolog G). | |||||
|
BATF_MOUSE
|
||||||
| NC score | 0.166286 (rank : 35) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.164506 (rank : 36) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.163951 (rank : 37) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MAFB_HUMAN
|
||||||
| NC score | 0.161410 (rank : 38) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
MAFF_MOUSE
|
||||||
| NC score | 0.160157 (rank : 39) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54791 | Gene names | Maff | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.158290 (rank : 40) | θ value | 0.125558 (rank : 33) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
MAFF_HUMAN
|
||||||
| NC score | 0.158176 (rank : 41) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ULX9, Q9Y525 | Gene names | MAFF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafF (V-maf musculoaponeurotic fibrosarcoma oncogene homolog F) (U-Maf). | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.157385 (rank : 42) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
BATF_HUMAN
|
||||||
| NC score | 0.155976 (rank : 43) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
NRL_HUMAN
|
||||||
| NC score | 0.155094 (rank : 44) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P54845 | Gene names | NRL | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL) (D14S46E). | |||||
|
NRL_MOUSE
|
||||||
| NC score | 0.154934 (rank : 45) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54846 | Gene names | Nrl | |||
|
Domain Architecture |

|
|||||
| Description | Neural retina-specific leucine zipper protein (NRL). | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.145703 (rank : 46) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.144207 (rank : 47) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.142888 (rank : 48) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.129011 (rank : 49) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.121291 (rank : 50) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
CEBPE_MOUSE
|
||||||
| NC score | 0.116445 (rank : 51) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
NFE2_HUMAN
|
||||||
| NC score | 0.115747 (rank : 52) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.111622 (rank : 53) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.110279 (rank : 54) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.100263 (rank : 55) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.097703 (rank : 56) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
NF2L3_MOUSE
|
||||||
| NC score | 0.096959 (rank : 57) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
CDC37_HUMAN
|
||||||
| NC score | 0.096411 (rank : 58) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.094944 (rank : 59) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
CEBPD_HUMAN
|
||||||
| NC score | 0.090578 (rank : 60) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.086613 (rank : 61) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
BACH1_HUMAN
|
||||||
| NC score | 0.084318 (rank : 62) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| NC score | 0.084275 (rank : 63) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.082607 (rank : 64) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CEBPB_MOUSE
|
||||||
| NC score | 0.076148 (rank : 65) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
CEBPD_MOUSE
|
||||||
| NC score | 0.074876 (rank : 66) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
BACH2_HUMAN
|
||||||
| NC score | 0.073855 (rank : 67) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH2_MOUSE
|
||||||
| NC score | 0.070615 (rank : 68) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.065683 (rank : 69) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.065683 (rank : 70) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.063126 (rank : 71) | θ value | 0.00134147 (rank : 16) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.062262 (rank : 72) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
CEBPA_MOUSE
|
||||||
| NC score | 0.061754 (rank : 73) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.061615 (rank : 74) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.058749 (rank : 75) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CEBPG_HUMAN
|
||||||
| NC score | 0.057348 (rank : 76) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
ATF5_MOUSE
|
||||||
| NC score | 0.056399 (rank : 77) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
CEBPG_MOUSE
|
||||||
| NC score | 0.056291 (rank : 78) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
VPS45_HUMAN
|
||||||
| NC score | 0.055647 (rank : 79) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRW7, Q15715, Q9Y4Z6 | Gene names | VPS45A, VPS45 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 45 (h-VPS45) (hlVps45). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.054779 (rank : 80) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.054604 (rank : 81) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.053811 (rank : 82) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.052509 (rank : 83) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
VPS45_MOUSE
|
||||||
| NC score | 0.051812 (rank : 84) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97390, Q91VK9 | Gene names | Vps45a, Vps45 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 45 (mVps45). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.051296 (rank : 85) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
ATF4_HUMAN
|
||||||
| NC score | 0.050454 (rank : 86) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.050086 (rank : 87) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.047929 (rank : 88) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.046986 (rank : 89) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.046818 (rank : 90) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
PHC1_MOUSE
|
||||||
| NC score | 0.045296 (rank : 91) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.044942 (rank : 92) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.044651 (rank : 93) | θ value | 0.0148317 (rank : 21) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.044403 (rank : 94) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.044387 (rank : 95) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.043084 (rank : 96) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
COVA1_MOUSE
|
||||||
| NC score | 0.042183 (rank : 97) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R0Z2, Q8BY59, Q8R372 | Gene names | Cova1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.041752 (rank : 98) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TEX15_HUMAN
|
||||||
| NC score | 0.041321 (rank : 99) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.040979 (rank : 100) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.040924 (rank : 101) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.040615 (rank : 102) | θ value | 0.62314 (rank : 58) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.040206 (rank : 103) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
NDE1_MOUSE
|
||||||
| NC score | 0.039921 (rank : 104) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CZA6, Q3UBS6, Q3UIC1, Q9ERR0 | Gene names | Nde1, Nude | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear distribution protein nudE homolog 1 (NudE) (mNudE). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.039889 (rank : 105) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.039660 (rank : 106) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.039563 (rank : 107) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
COVA1_HUMAN
|
||||||
| NC score | 0.039414 (rank : 108) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q16206, Q5VTJ1, Q5VTJ2, Q8WUX0, Q9NTP6, Q9UH82 | Gene names | COVA1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated hydroquinone oxidase (tNOX) (Cytosolic ovarian carcinoma antigen 1) (APK1 antigen) [Includes: Hydroquinone [NADH] oxidase (EC 1.-.-.-); Protein disulfide-thiol oxidoreductase (EC 1.-.-.-)]. | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.039163 (rank : 109) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.038721 (rank : 110) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.038713 (rank : 111) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.038635 (rank : 112) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.038560 (rank : 113) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.037551 (rank : 114) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.037196 (rank : 115) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.037162 (rank : 116) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.036966 (rank : 117) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.036956 (rank : 118) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.036869 (rank : 119) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.036477 (rank : 120) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
POF1B_HUMAN
|
||||||
| NC score | 0.036117 (rank : 121) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.035538 (rank : 122) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.035001 (rank : 123) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.034584 (rank : 124) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
PLVAP_HUMAN
|
||||||
| NC score | 0.034024 (rank : 125) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BX97, Q86VP0, Q8N8Y0, Q8ND68, Q8TER8, Q9BZD5 | Gene names | PLVAP, FELS, PV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plasmalemma vesicle-associated protein (Plasmalemma vesicle protein 1) (PV-1) (Fenestrated endothelial-linked structure protein). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.033903 (rank : 126) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.032526 (rank : 127) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.032279 (rank : 128) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.031002 (rank : 129) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.030442 (rank : 130) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.027936 (rank : 131) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
HOME2_HUMAN
|
||||||
| NC score | 0.027784 (rank : 132) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NSB8, O95269, O95349, Q9NSB6, Q9NSB7, Q9UNT7 | Gene names | HOMER2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin). | |||||
|
K1377_HUMAN
|
||||||
| NC score | 0.027469 (rank : 133) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2H0, Q4G0U6 | Gene names | KIAA1377 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1377. | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.027219 (rank : 134) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.027034 (rank : 135) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.026968 (rank : 136) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.026930 (rank : 137) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.026466 (rank : 138) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.026213 (rank : 139) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.025843 (rank : 140) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.025392 (rank : 141) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
FBXL3_MOUSE
|
||||||
| NC score | 0.025382 (rank : 142) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4V4, Q80V32, Q8K1W0, Q9QXW1 | Gene names | Fbxl3, Fbl3a, Fbxl3a | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 3 (F-box/LRR-repeat protein 3A) (F-box and leucine-rich repeat protein 3A). | |||||
|
FBXL3_HUMAN
|
||||||
| NC score | 0.025351 (rank : 143) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKT7, Q9P122 | Gene names | FBXL3, FBL3A, FBXL3A | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 3 (F-box/LRR-repeat protein 3A) (F-box and leucine-rich repeat protein 3A). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.025252 (rank : 144) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.025018 (rank : 145) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.024798 (rank : 146) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.023917 (rank : 147) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.023871 (rank : 148) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.022960 (rank : 149) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.022870 (rank : 150) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.022473 (rank : 151) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.022434 (rank : 152) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
THAP4_HUMAN
|
||||||
| NC score | 0.022218 (rank : 153) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WY91, Q53NU7, Q6GRN0, Q6IPJ3, Q9NW26, Q9Y325 | Gene names | THAP4 | |||
|
Domain Architecture |

|
|||||
| Description | THAP domain-containing protein 4. | |||||
|
DAAM1_MOUSE
|
||||||
| NC score | 0.021503 (rank : 154) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BPM0, Q80Y68, Q9CQQ2 | Gene names | Daam1 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.021456 (rank : 155) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
BORG5_MOUSE
|
||||||
| NC score | 0.021139 (rank : 156) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W92, Q9D8M1 | Gene names | Cdc42ep1, Borg5, Cep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.020282 (rank : 157) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.019808 (rank : 158) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.019625 (rank : 159) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.019406 (rank : 160) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MCA1_HUMAN
|
||||||
| NC score | 0.019291 (rank : 161) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12904, Q6FG28, Q96CQ9 | Gene names | SCYE1, EMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
MOR2A_MOUSE
|
||||||
| NC score | 0.019156 (rank : 162) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
G3BP_HUMAN
|
||||||
| NC score | 0.018312 (rank : 163) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13283 | Gene names | G3BP, G3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.018263 (rank : 164) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.018135 (rank : 165) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.017263 (rank : 166) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.017235 (rank : 167) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
VP33B_HUMAN
|
||||||
| NC score | 0.017033 (rank : 168) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H267, Q96K14, Q9NRP6, Q9NSF3 | Gene names | VPS33B | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 33B (hVPS33B). | |||||
|
VP33B_MOUSE
|
||||||
| NC score | 0.017009 (rank : 169) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59016 | Gene names | Vps33b | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 33B. | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.016658 (rank : 170) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
OTU6A_HUMAN
|
||||||
| NC score | 0.016277 (rank : 171) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L8S5 | Gene names | OTUD6A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 6A. | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.015925 (rank : 172) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
KI18A_HUMAN
|
||||||
| NC score | 0.015454 (rank : 173) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NI77, Q86VS5, Q9H0F3 | Gene names | KIF18A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 18A. | |||||
|
MRCKG_MOUSE
|
||||||
| NC score | 0.015364 (rank : 174) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ZHX1_HUMAN
|
||||||
| NC score | 0.014971 (rank : 175) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
FHOD1_HUMAN
|
||||||
| NC score | 0.014154 (rank : 176) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.013349 (rank : 177) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.011256 (rank : 178) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.010929 (rank : 179) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.010134 (rank : 180) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
L2GL2_MOUSE
|
||||||
| NC score | 0.009851 (rank : 181) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TJ91, Q6YP20, Q8K1X0 | Gene names | Llgl2, Llglh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(2) giant larvae protein homolog 2 (Lethal giant larvae-like protein 2). | |||||
|
LRC59_HUMAN
|
||||||
| NC score | 0.009391 (rank : 182) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96AG4, Q9P189 | Gene names | LRRC59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 59. | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.008879 (rank : 183) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
VDR_HUMAN
|
||||||
| NC score | 0.008322 (rank : 184) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11473 | Gene names | VDR, NR1I1 | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
GEMI5_MOUSE
|
||||||
| NC score | 0.006470 (rank : 185) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.006150 (rank : 186) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MYLK_HUMAN
|
||||||
| NC score | 0.003458 (rank : 187) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15746, O95796, O95797, O95798, O95799, Q14844, Q16794, Q5MY99, Q5MYA0, Q7Z4J0, Q9C0L5, Q9UBG5, Q9UIT9 | Gene names | MYLK, MLCK | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
ZN509_HUMAN
|
||||||
| NC score | -0.002774 (rank : 188) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||