Please be patient as the page loads
|
FOSB_MOUSE
|
||||||
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FOSB_MOUSE
|
||||||
| θ value | 5.41814e-138 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 4.44261e-132 (rank : 2) | NC score | 0.990085 (rank : 2) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOS_MOUSE
|
||||||
| θ value | 2.67802e-36 (rank : 3) | NC score | 0.878153 (rank : 3) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
FOS_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 4) | NC score | 0.873669 (rank : 4) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | 1.57365e-28 (rank : 5) | NC score | 0.866984 (rank : 5) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_HUMAN
|
||||||
| θ value | 3.50572e-28 (rank : 6) | NC score | 0.866764 (rank : 6) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 7) | NC score | 0.800428 (rank : 7) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 8) | NC score | 0.798929 (rank : 8) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
ATF3_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 9) | NC score | 0.650445 (rank : 10) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
ATF3_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 10) | NC score | 0.647562 (rank : 12) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
BATF_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 11) | NC score | 0.658845 (rank : 9) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |
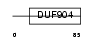
|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
JUNB_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 12) | NC score | 0.449627 (rank : 13) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 13) | NC score | 0.445658 (rank : 16) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 14) | NC score | 0.446637 (rank : 15) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
BATF_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 15) | NC score | 0.649092 (rank : 11) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |
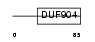
|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
JUNB_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 16) | NC score | 0.439999 (rank : 17) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |
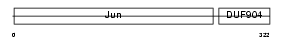
|
|||||
| Description | Transcription factor jun-B. | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 17) | NC score | 0.437236 (rank : 18) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 18) | NC score | 0.436480 (rank : 19) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
JUND_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 19) | NC score | 0.428834 (rank : 20) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 20) | NC score | 0.423371 (rank : 21) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUN_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 21) | NC score | 0.405722 (rank : 23) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
JUN_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 22) | NC score | 0.406368 (rank : 22) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 23) | NC score | 0.449064 (rank : 14) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 24) | NC score | 0.078261 (rank : 62) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
TEF_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 25) | NC score | 0.277988 (rank : 27) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TEF_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 26) | NC score | 0.278481 (rank : 26) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
CEBPG_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 27) | NC score | 0.307982 (rank : 25) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |
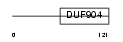
|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
EPN3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.077761 (rank : 63) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.078341 (rank : 61) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CEBPG_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 30) | NC score | 0.308813 (rank : 24) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
CDC37_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.132830 (rank : 48) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 32) | NC score | 0.071940 (rank : 68) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.093097 (rank : 56) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.158290 (rank : 43) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.255239 (rank : 28) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.075838 (rank : 65) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
ATF5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.176548 (rank : 39) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.199192 (rank : 36) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.198154 (rank : 37) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.032525 (rank : 167) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.219469 (rank : 33) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.030097 (rank : 172) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.065621 (rank : 76) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.053665 (rank : 113) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.165562 (rank : 41) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
CEBPA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.224007 (rank : 32) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.225134 (rank : 31) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.051070 (rank : 136) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.077554 (rank : 64) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
ARRD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.033562 (rank : 163) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
DBP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.171495 (rank : 40) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
DBP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.185392 (rank : 38) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.072461 (rank : 67) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
USBP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.085188 (rank : 59) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8N6Y0, Q8NBX7, Q96KH3, Q9BYI8 | Gene names | USHBP1, AIEBP, MCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1) (MCC-2) (AIE-75 binding protein). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.032672 (rank : 166) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.024356 (rank : 178) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 57) | NC score | 0.162399 (rank : 42) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
BACH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.123508 (rank : 49) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.123037 (rank : 50) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.071502 (rank : 70) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CRCM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.084597 (rank : 60) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |
|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
KIF12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.028810 (rank : 173) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96FN5, Q5TBE0 | Gene names | KIF12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF12. | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.037282 (rank : 158) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
CEBPD_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.238563 (rank : 29) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
CEBPD_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.230416 (rank : 30) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
CLCB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.032701 (rank : 165) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
GLI3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.008480 (rank : 190) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.054581 (rank : 102) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.068936 (rank : 74) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.054994 (rank : 100) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.052239 (rank : 123) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DMN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.027695 (rank : 174) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.058553 (rank : 88) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.068546 (rank : 75) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
NF2L3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.152531 (rank : 45) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.017386 (rank : 182) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.013475 (rank : 188) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.050752 (rank : 142) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.037061 (rank : 159) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.069225 (rank : 73) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CEBPB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.211635 (rank : 34) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.042295 (rank : 153) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.030671 (rank : 170) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.033722 (rank : 161) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.060571 (rank : 82) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.056127 (rank : 95) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.056434 (rank : 93) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
IDLC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.073649 (rank : 66) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BVN8 | Gene names | Dnali1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal). | |||||
|
M3K7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.002146 (rank : 195) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62073 | Gene names | Map3k7, Tak1 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||
|
MAP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.026721 (rank : 175) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.004894 (rank : 193) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.049031 (rank : 147) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.024824 (rank : 177) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.023960 (rank : 179) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.030565 (rank : 171) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
AZI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.061618 (rank : 80) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.059950 (rank : 84) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.048761 (rank : 148) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.056605 (rank : 91) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CREB3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.150773 (rank : 46) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.054285 (rank : 106) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.005855 (rank : 191) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.055104 (rank : 98) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.051809 (rank : 125) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.030716 (rank : 169) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
XBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.206440 (rank : 35) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
DBNL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.015048 (rank : 186) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.055008 (rank : 99) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.037645 (rank : 157) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.038568 (rank : 156) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.052534 (rank : 120) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.016666 (rank : 184) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.046840 (rank : 150) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BAP29_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.042122 (rank : 154) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UHQ4, O95003 | Gene names | BCAP29, BAP29 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 29 (BCR-associated protein Bap29). | |||||
|
CEBPE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.154803 (rank : 44) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.050954 (rank : 137) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.002975 (rank : 194) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.025480 (rank : 176) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.047364 (rank : 149) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.057066 (rank : 89) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PACN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.017293 (rank : 183) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.056325 (rank : 94) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.005625 (rank : 192) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
SETX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.019681 (rank : 180) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.013617 (rank : 187) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.012272 (rank : 189) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
ZCHC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.033632 (rank : 162) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8U3 | Gene names | ZCCHC5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCHC domain-containing protein 5. | |||||
|
ADIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.045750 (rank : 151) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
DIAP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.018392 (rank : 181) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
KI21B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.033901 (rank : 160) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
KI21B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.033409 (rank : 164) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
KIF23_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.039936 (rank : 155) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
M3K7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.001296 (rank : 196) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43318, O43317, O43319, Q5TDN2, Q5TDN3 | Gene names | MAP3K7, TAK1 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.051441 (rank : 129) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
TNR8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.016483 (rank : 185) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.030937 (rank : 168) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.043781 (rank : 152) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
ATF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.071204 (rank : 71) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.069656 (rank : 72) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
ATF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.086749 (rank : 58) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
ATF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.090673 (rank : 57) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
ATF5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.071643 (rank : 69) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.054453 (rank : 104) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
BACH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.054635 (rank : 101) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.054406 (rank : 105) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.055945 (rank : 96) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.053667 (rank : 112) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CCD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.056676 (rank : 90) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051117 (rank : 135) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
CDC37_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.062318 (rank : 79) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |
|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.054010 (rank : 108) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CING_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050779 (rank : 141) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CN145_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050914 (rank : 139) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CP135_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.050933 (rank : 138) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052088 (rank : 124) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CREB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.059216 (rank : 87) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.059234 (rank : 86) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |
|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.104510 (rank : 55) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
CREM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.063832 (rank : 77) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |
|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.063455 (rank : 78) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050510 (rank : 144) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DDIT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.056532 (rank : 92) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
DDIT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.054474 (rank : 103) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.051714 (rank : 126) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052424 (rank : 121) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
GOG8A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.051257 (rank : 132) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.053543 (rank : 114) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.053965 (rank : 109) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
HLF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.106935 (rank : 52) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
HLF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.106687 (rank : 53) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.052797 (rank : 118) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.052296 (rank : 122) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.052770 (rank : 119) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
HOOK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.053372 (rank : 116) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
IDLC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.059520 (rank : 85) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14645, Q5TGH0, Q7L0I5 | Gene names | DNALI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal) (hp28). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.051272 (rank : 131) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.050795 (rank : 140) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
INCE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.051190 (rank : 134) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.055737 (rank : 97) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.053933 (rank : 111) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.050353 (rank : 146) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.053941 (rank : 110) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.110809 (rank : 51) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
NFE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.106637 (rank : 54) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.053395 (rank : 115) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.051304 (rank : 130) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.051703 (rank : 127) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.050483 (rank : 145) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RFIP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.060371 (rank : 83) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
RFIP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.051652 (rank : 128) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.050748 (rank : 143) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
TMF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.054135 (rank : 107) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TNIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.051200 (rank : 133) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.052938 (rank : 117) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
USBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.060724 (rank : 81) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R370 | Gene names | Ushbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1). | |||||
|
XBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.139544 (rank : 47) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
FOSB_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.41814e-138 (rank : 1) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | P13346 | Gene names | Fosb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB. | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.990085 (rank : 2) | θ value | 4.44261e-132 (rank : 2) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
FOS_MOUSE
|
||||||
| NC score | 0.878153 (rank : 3) | θ value | 2.67802e-36 (rank : 3) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P01101 | Gene names | Fos | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos). | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.873669 (rank : 4) | θ value | 5.96599e-36 (rank : 4) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.866984 (rank : 5) | θ value | 1.57365e-28 (rank : 5) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL2_HUMAN
|
||||||
| NC score | 0.866764 (rank : 6) | θ value | 3.50572e-28 (rank : 6) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P15408 | Gene names | FOSL2, FRA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.800428 (rank : 7) | θ value | 2.60593e-15 (rank : 7) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.798929 (rank : 8) | θ value | 5.8054e-15 (rank : 8) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
BATF_MOUSE
|
||||||
| NC score | 0.658845 (rank : 9) | θ value | 1.17247e-07 (rank : 11) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35284 | Gene names | Batf | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcription factor B-ATF. | |||||
|
ATF3_HUMAN
|
||||||
| NC score | 0.650445 (rank : 10) | θ value | 6.87365e-08 (rank : 9) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P18847 | Gene names | ATF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3). | |||||
|
BATF_HUMAN
|
||||||
| NC score | 0.649092 (rank : 11) | θ value | 2.21117e-06 (rank : 15) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16520 | Gene names | BATF | |||
|
Domain Architecture |

|
|||||
| Description | ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT- activated gene 2) (SFA-2). | |||||
|
ATF3_MOUSE
|
||||||
| NC score | 0.647562 (rank : 12) | θ value | 6.87365e-08 (rank : 10) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q60765 | Gene names | Atf3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-3 (Activating transcription factor 3) (Transcription factor LRG-21). | |||||
|
JUNB_HUMAN
|
||||||
| NC score | 0.449627 (rank : 13) | θ value | 4.45536e-07 (rank : 12) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P17275, Q96GH3 | Gene names | JUNB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.449064 (rank : 14) | θ value | 0.000786445 (rank : 23) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.446637 (rank : 15) | θ value | 1.29631e-06 (rank : 14) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.445658 (rank : 16) | θ value | 1.29631e-06 (rank : 13) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
JUNB_MOUSE
|
||||||
| NC score | 0.439999 (rank : 17) | θ value | 3.77169e-06 (rank : 16) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P09450, Q8C2G9 | Gene names | Junb, Jun-b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-B. | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.437236 (rank : 18) | θ value | 1.87187e-05 (rank : 17) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.436480 (rank : 19) | θ value | 3.19293e-05 (rank : 18) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
JUND_HUMAN
|
||||||
| NC score | 0.428834 (rank : 20) | θ value | 0.00020696 (rank : 19) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P17535 | Gene names | JUND | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.423371 (rank : 21) | θ value | 0.00020696 (rank : 20) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
JUN_MOUSE
|
||||||
| NC score | 0.406368 (rank : 22) | θ value | 0.000461057 (rank : 22) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05627 | Gene names | Jun | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (Jun A) (AH119). | |||||
|
JUN_HUMAN
|
||||||
| NC score | 0.405722 (rank : 23) | θ value | 0.000461057 (rank : 21) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05412, Q96G93 | Gene names | JUN | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-1 (Activator protein 1) (AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus 17 oncogene homolog) (p39). | |||||
|
CEBPG_MOUSE
|
||||||
| NC score | 0.308813 (rank : 24) | θ value | 0.0193708 (rank : 30) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P53568 | Gene names | Cebpg | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma) (Immunoglobulin enhancer-binding protein 1) (IG/EBP-1) (Granulocyte colony-stimulating factor promoter element 1-binding protein) (GPE1-binding protein) (GPE1-BP). | |||||
|
CEBPG_HUMAN
|
||||||
| NC score | 0.307982 (rank : 25) | θ value | 0.0148317 (rank : 27) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53567, Q5U052 | Gene names | CEBPG | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein gamma (C/EBP gamma). | |||||
|
TEF_MOUSE
|
||||||
| NC score | 0.278481 (rank : 26) | θ value | 0.00298849 (rank : 26) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
TEF_HUMAN
|
||||||
| NC score | 0.277988 (rank : 27) | θ value | 0.00298849 (rank : 25) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q10587, Q15729, Q7Z3J7, Q8IU94, Q96TG4 | Gene names | TEF | |||
|
Domain Architecture |

|
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.255239 (rank : 28) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
CEBPD_HUMAN
|
||||||
| NC score | 0.238563 (rank : 29) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
CEBPD_MOUSE
|
||||||
| NC score | 0.230416 (rank : 30) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
CEBPA_MOUSE
|
||||||
| NC score | 0.225134 (rank : 31) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.224007 (rank : 32) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.219469 (rank : 33) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CEBPB_MOUSE
|
||||||
| NC score | 0.211635 (rank : 34) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.206440 (rank : 35) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.199192 (rank : 36) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.198154 (rank : 37) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
DBP_MOUSE
|
||||||
| NC score | 0.185392 (rank : 38) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
ATF5_HUMAN
|
||||||
| NC score | 0.176548 (rank : 39) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
DBP_HUMAN
|
||||||
| NC score | 0.171495 (rank : 40) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.165562 (rank : 41) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.162399 (rank : 42) | θ value | 0.62314 (rank : 57) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.158290 (rank : 43) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
CEBPE_MOUSE
|
||||||
| NC score | 0.154803 (rank : 44) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PZD9 | Gene names | Cebpe | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
NF2L3_MOUSE
|
||||||
| NC score | 0.152531 (rank : 45) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
CREB3_MOUSE
|
||||||
| NC score | 0.150773 (rank : 46) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61817, Q99M21, Q9CVK9 | Gene names | Creb3, Lzip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Transcription factor LZIP). | |||||
|
XBP1_HUMAN
|
||||||
| NC score | 0.139544 (rank : 47) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17861, Q8WYK6, Q969P1, Q96BD7 | Gene names | XBP1, TREB5, XBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5). | |||||
|
CDC37_HUMAN
|
||||||
| NC score | 0.132830 (rank : 48) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q16543 | Gene names | CDC37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
BACH1_HUMAN
|
||||||
| NC score | 0.123508 (rank : 49) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| NC score | 0.123037 (rank : 50) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.110809 (rank : 51) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
HLF_HUMAN
|
||||||
| NC score | 0.106935 (rank : 52) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q16534 | Gene names | HLF | |||
|
Domain Architecture |

|
|||||
| Description | Hepatic leukemia factor. | |||||
|
HLF_MOUSE
|
||||||
| NC score | 0.106687 (rank : 53) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BW74, Q6PF83 | Gene names | Hlf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatic leukemia factor. | |||||
|
NFE2_HUMAN
|
||||||
| NC score | 0.106637 (rank : 54) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16621, Q07720, Q6ICV9 | Gene names | NFE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor NF-E2 45 kDa subunit (Nuclear factor, erythroid- derived 2 45 kDa subunit) (p45 NF-E2) (Leucine zipper protein NF-E2). | |||||
|
CREB3_HUMAN
|
||||||
| NC score | 0.104510 (rank : 55) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43889, O14671, O14919, Q96GK8, Q9H2W3, Q9UE77 | Gene names | CREB3, LZIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-responsive element-binding protein 3 (Luman protein) (Transcription factor LZIP-alpha). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.093097 (rank : 56) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
ATF4_MOUSE
|
||||||
| NC score | 0.090673 (rank : 57) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q06507, Q61906 | Gene names | Atf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (C/EBP-related ATF) (C/ATF) (TAXREB67 homolog). | |||||
|
ATF4_HUMAN
|
||||||
| NC score | 0.086749 (rank : 58) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P18848, Q9UH31 | Gene names | ATF4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-4 (Activating transcription factor 4) (DNA-binding protein TAXREB67) (Cyclic AMP response element-binding protein 2) (CREB2). | |||||
|
USBP1_HUMAN
|
||||||
| NC score | 0.085188 (rank : 59) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8N6Y0, Q8NBX7, Q96KH3, Q9BYI8 | Gene names | USHBP1, AIEBP, MCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1) (MCC-2) (AIE-75 binding protein). | |||||
|
CRCM_HUMAN
|
||||||
| NC score | 0.084597 (rank : 60) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |

|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.078341 (rank : 61) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.078261 (rank : 62) | θ value | 0.000786445 (rank : 24) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
EPN3_HUMAN
|
||||||
| NC score | 0.077761 (rank : 63) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H201, Q9BVN6, Q9NWK2 | Gene names | EPN3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.077554 (rank : 64) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.075838 (rank : 65) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
IDLC_MOUSE
|
||||||
| NC score | 0.073649 (rank : 66) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BVN8 | Gene names | Dnali1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.072461 (rank : 67) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.071940 (rank : 68) | θ value | 0.0563607 (rank : 32) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ATF5_MOUSE
|
||||||
| NC score | 0.071643 (rank : 69) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.071502 (rank : 70) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
ATF1_HUMAN
|
||||||
| NC score | 0.071204 (rank : 71) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
ATF1_MOUSE
|
||||||
| NC score | 0.069656 (rank : 72) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.069225 (rank : 73) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.068936 (rank : 74) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.068546 (rank : 75) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.065621 (rank : 76) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CREM_HUMAN
|
||||||
| NC score | 0.063832 (rank : 77) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03060, Q16114, Q16116, Q9NZB9 | Gene names | CREM | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CREM_MOUSE
|
||||||
| NC score | 0.063455 (rank : 78) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
CDC37_MOUSE
|
||||||
| NC score | 0.062318 (rank : 79) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
AZI1_HUMAN
|
||||||
| NC score | 0.061618 (rank : 80) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1000 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9UPN4, Q96F50 | Gene names | AZI1, CEP131, KIAA1118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1) (Centrosomal protein of 131 kDa) (Cep131 protein). | |||||
|
USBP1_MOUSE
|
||||||
| NC score | 0.060724 (rank : 81) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R370 | Gene names | Ushbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.060571 (rank : 82) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
RFIP4_HUMAN
|
||||||
| NC score | 0.060371 (rank : 83) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.059950 (rank : 84) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
IDLC_HUMAN
|
||||||
| NC score | 0.059520 (rank : 85) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14645, Q5TGH0, Q7L0I5 | Gene names | DNALI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axonemal dynein light intermediate polypeptide 1 (Inner dynein arm light chain, axonemal) (hp28). | |||||
|
CREB1_MOUSE
|
||||||
| NC score | 0.059234 (rank : 86) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01147 | Gene names | Creb1, Creb-1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
CREB1_HUMAN
|
||||||
| NC score | 0.059216 (rank : 87) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16220, P21934, Q9UMA7 | Gene names | CREB1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein (CREB). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.058553 (rank : 88) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.057066 (rank : 89) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.056676 (rank : 90) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.056605 (rank : 91) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DDIT3_HUMAN
|
||||||
| NC score | 0.056532 (rank : 92) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.056434 (rank : 93) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.056325 (rank : 94) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.056127 (rank : 95) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.055945 (rank : 96) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.055737 (rank : 97) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.055104 (rank : 98) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.055008 (rank : 99) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.054994 (rank : 100) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BACH2_HUMAN
|
||||||
| NC score | 0.054635 (rank : 101) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.054581 (rank : 102) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
DDIT3_MOUSE
|
||||||
| NC score | 0.054474 (rank : 103) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.054453 (rank : 104) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
BACH2_MOUSE
|
||||||
| NC score | 0.054406 (rank : 105) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.054285 (rank : 106) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.054135 (rank : 107) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.054010 (rank : 108) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.053965 (rank : 109) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.053941 (rank : 110) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.053933 (rank : 111) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.053667 (rank : 112) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.053665 (rank : 113) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.053543 (rank : 114) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.053395 (rank : 115) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
HOOK2_MOUSE
|
||||||
| NC score | 0.053372 (rank : 116) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.052938 (rank : 117) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.052797 (rank : 118) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.052770 (rank : 119) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.052534 (rank : 120) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.052424 (rank : 121) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.052296 (rank : 122) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.052239 (rank : 123) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.052088 (rank : 124) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.051809 (rank : 125) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.051714 (rank : 126) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.051703 (rank : 127) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
RFIP4_MOUSE
|
||||||
| NC score | 0.051652 (rank : 128) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.051441 (rank : 129) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.051304 (rank : 130) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.051272 (rank : 131) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
GOG8A_HUMAN
|
||||||
| NC score | 0.051257 (rank : 132) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
TNIP3_HUMAN
|
||||||
| NC score | 0.051200 (rank : 133) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 511 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96KP6, Q96PQ3, Q9H780 | Gene names | TNIP3, ABIN3, LIND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TNFAIP3-interacting protein 3 (Listeria-induced gene protein) (A20- binding inhibitor of NF-kappa-B activation 3) (ABIN-3). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.051190 (rank : 134) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.051117 (rank : 135) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.051070 (rank : 136) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.050954 (rank : 137) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.050933 (rank : 138) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.050914 (rank : 139) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.050795 (rank : 140) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.050779 (rank : 141) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.050752 (rank : 142) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.050748 (rank : 143) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.050510 (rank : 144) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.050483 (rank : 145) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.050353 (rank : 146) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.049031 (rank : 147) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.048761 (rank : 148) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.047364 (rank : 149) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.046840 (rank : 150) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ADIP_MOUSE
|
||||||
| NC score | 0.045750 (rank : 151) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.043781 (rank : 152) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.042295 (rank : 153) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
BAP29_HUMAN
|
||||||
| NC score | 0.042122 (rank : 154) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UHQ4, O95003 | Gene names | BCAP29, BAP29 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 29 (BCR-associated protein Bap29). | |||||
|
KIF23_HUMAN
|
||||||
| NC score | 0.039936 (rank : 155) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.038568 (rank : 156) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.037645 (rank : 157) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.037282 (rank : 158) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.037061 (rank : 159) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
KI21B_HUMAN
|
||||||
| NC score | 0.033901 (rank : 160) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1017 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75037, Q5T4J3 | Gene names | KIF21B, KIAA0449 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B. | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.033722 (rank : 161) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
ZCHC5_HUMAN
|
||||||
| NC score | 0.033632 (rank : 162) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N8U3 | Gene names | ZCCHC5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCHC domain-containing protein 5. | |||||
|
ARRD1_HUMAN
|
||||||
| NC score | 0.033562 (rank : 163) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
KI21B_MOUSE
|
||||||
| NC score | 0.033409 (rank : 164) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
CLCB_MOUSE
|
||||||
| NC score | 0.032701 (rank : 165) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IRU5, Q8BR04, Q8CDX9, Q9CV35 | Gene names | Cltb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.032672 (rank : 166) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.032525 (rank : 167) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.030937 (rank : 168) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.030716 (rank : 169) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.030671 (rank : 170) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.030565 (rank : 171) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.030097 (rank : 172) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
KIF12_HUMAN
|
||||||
| NC score | 0.028810 (rank : 173) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96FN5, Q5TBE0 | Gene names | KIF12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF12. | |||||
|
DMN_HUMAN
|
||||||
| NC score | 0.027695 (rank : 174) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15061 | Gene names | DMN, KIAA0353 | |||
|
Domain Architecture |

|
|||||
| Description | Desmuslin. | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.026721 (rank : 175) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.025480 (rank : 176) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.024824 (rank : 177) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.024356 (rank : 178) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.023960 (rank : 179) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.019681 (rank : 180) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
DIAP3_MOUSE
|
||||||
| NC score | 0.018392 (rank : 181) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.017386 (rank : 182) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PACN3_HUMAN
|
||||||
| NC score | 0.017293 (rank : 183) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.016666 (rank : 184) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.016483 (rank : 185) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
DBNL_MOUSE
|
||||||
| NC score | 0.015048 (rank : 186) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.013617 (rank : 187) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.013475 (rank : 188) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.012272 (rank : 189) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
GLI3_HUMAN
|
||||||
| NC score | 0.008480 (rank : 190) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10071, O75219, Q9UDT5, Q9UJ39 | Gene names | GLI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
GLI3_MOUSE
|
||||||
| NC score | 0.005855 (rank : 191) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.005625 (rank : 192) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.004894 (rank : 193) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.002975 (rank : 194) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
M3K7_MOUSE
|
||||||
| NC score | 0.002146 (rank : 195) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62073 | Gene names | Map3k7, Tak1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||
|
M3K7_HUMAN
|
||||||
| NC score | 0.001296 (rank : 196) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 137 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43318, O43317, O43319, Q5TDN2, Q5TDN3 | Gene names | MAP3K7, TAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||